 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Neem_8508_f_1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Meliaceae; Azadirachta
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 764aa MW: 82227.8 Da PI: 6.4223 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 50.6 | 3.5e-16 | 476 | 522 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K++Ka++L +A+eY+k Lq
Neem_8508_f_1 476 VHNLSERRRRDRINEKMRALQELIPNC-----NKVDKASMLDEAIEYLKTLQ 522
6*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 2.81E-17 | 468 | 526 | No hit | No description |
| SuperFamily | SSF47459 | 1.83E-20 | 469 | 532 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 2.8E-20 | 469 | 530 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.349 | 472 | 521 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.2E-13 | 476 | 522 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.7E-17 | 478 | 527 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 764 aa Download sequence Send to blast |
MPLFELYRMT KEKLDSAKQN NSSPSNDPSF QPENDFVELV WENGPILMQG QSSKARKSPT 60 SNNLSSQCVP SHTTKTRDKD VGNGTKVGKF GTVDSVLNEI PMSVPSGEMG LNQDDEMLRW 120 LNYSVDESLE HEYCSDFLPE LSGITGNELS SQNNFSSFDK RSSSSNQFGK EFKTVCVHND 180 LSLEQGNLSK ISSVGGGDGT RPKGSASQLY TSSSQQYQTS FPYFRSRVSD STGSSVKNSA 240 ENAMAGGSNP VSGGSLSGVK IQKQDQRISS NKSGFMNFSH FTRPVALAKA NLQNLGSMSG 300 SGLSRIERMG SKDKSSVASS SNPVDPTPIH SSVGLRKEVS SQCQPVAGPS KVDIKPLDVK 360 LPELAAAAEQ PEVRCQEDVC NNDKRPSQNL GESATKELLD SEKTVEPVVA SSSVCSGNSV 420 EPASDDHPTH NLKRKHRDNE DSECPSEDVE EESVVVKKAV PTRVGSGSKR SRAAEVHNLS 480 ERRRRDRINE KMRALQELIP NCNKVDKASM LDEAIEYLKT LQLQVQIMSM GAGLYVPPMM 540 FQPGMQHMHP AAHMAPYSPM GMGMGMGIGF GMGMPDMNGS SSCYPMFQVP PLHGAHFPSP 600 SVSGPAVMQA MAGSNFPLFG LHGQGHPMSM PHTPLTALSG GPMMKSAMGL NACGMVGPMD 660 NFDAATASTS KDQVQNPNLQ VMPKTGASSS VNPTSGQVSK FIMQSHFRKV VDSSGEWECQ 720 TTNEGFAQPT GVQENCQAAA VNGRGTFNSS SKNDDESNRQ ACYD |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 480 | 485 | ERRRRD |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
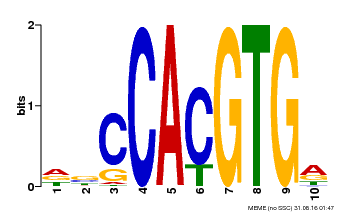
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006423962.1 | 0.0 | transcription factor PIF3 | ||||
| Refseq | XP_006480339.1 | 0.0 | transcription factor PIF3 | ||||
| TrEMBL | V4SJJ6 | 0.0 | V4SJJ6_9ROSI; Uncharacterized protein | ||||
| STRING | XP_006480339.1 | 0.0 | (Citrus sinensis) | ||||
| STRING | XP_006423961.1 | 0.0 | (Citrus clementina) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM7665 | 27 | 39 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 1e-50 | phytochrome interacting factor 3 | ||||



