 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Niben101Scf00061g00002.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Nicotianoideae; Nicotianeae; Nicotiana
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 272aa MW: 30718.2 Da PI: 9.0029 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 99.9 | 9.7e-32 | 9 | 58 | 1 | 50 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeys 50
krienk+nrqvtfskRrng+lKKA+ELSvLCdaeva+iifss+gklye+
Niben101Scf00061g00002.1 9 KRIENKINRQVTFSKRRNGLLKKAYELSVLCDAEVALIIFSSRGKLYEFG 58
79**********************************************95 PP
| |||||||
| 2 | K-box | 102.3 | 6.3e-34 | 80 | 169 | 9 | 98 |
K-box 9 leeakaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelqeenka 93
e++++s++qe++kLk++ e Lqr+qRhllGedL++LslkeLq+Le+qLe +l + R++K++++ eq+eel++ke++l + nk+
Niben101Scf00061g00002.1 80 CGERETQSWYQEVSKLKAKFEALQRTQRHLLGEDLGPLSLKELQSLEKQLEGALAQARQRKTQIMAEQMEELRRKERQLGDVNKQ 164
67899******************************************************************************** PP
K-box 94 Lrkkl 98
L+ k+
Niben101Scf00061g00002.1 165 LKIKV 169
*9886 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00432 | 3.5E-42 | 1 | 60 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS50066 | 33.683 | 1 | 61 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 2.08E-44 | 2 | 72 | No hit | No description |
| SuperFamily | SSF55455 | 1.96E-33 | 2 | 77 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 8.9E-33 | 3 | 23 | IPR002100 | Transcription factor, MADS-box |
| PROSITE pattern | PS00350 | 0 | 3 | 57 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 3.4E-27 | 10 | 57 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 8.9E-33 | 23 | 38 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 8.9E-33 | 38 | 59 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF01486 | 8.6E-31 | 82 | 169 | IPR002487 | Transcription factor, K-box |
| PROSITE profile | PS51297 | 16.592 | 85 | 179 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009553 | Biological Process | embryo sac development | ||||
| GO:0009911 | Biological Process | positive regulation of flower development | ||||
| GO:0010094 | Biological Process | specification of carpel identity | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0010582 | Biological Process | floral meristem determinacy | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0048455 | Biological Process | stamen formation | ||||
| GO:0048459 | Biological Process | floral whorl structural organization | ||||
| GO:0048509 | Biological Process | regulation of meristem development | ||||
| GO:0048833 | Biological Process | specification of floral organ number | ||||
| GO:0080060 | Biological Process | integument development | ||||
| GO:0080112 | Biological Process | seed growth | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 272 aa Download sequence Send to blast |
MGRGRVELKR IENKINRQVT FSKRRNGLLK KAYELSVLCD AEVALIIFSS RGKLYEFGSS 60 GITKTLERYQ RCCLNPQDNC GERETQSWYQ EVSKLKAKFE ALQRTQRHLL GEDLGPLSLK 120 ELQSLEKQLE GALAQARQRK TQIMAEQMEE LRRKERQLGD VNKQLKIKVS LELSSLEAEG 180 QGLRALPFPW TCNASAGSSS FAVHPSQSNH MECEPDPVLQ IGYHHYMAAE GASGSRSMAV 240 ESIITIWLQK EPQGQGAWLL RVISSMVGAF DF |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ox0_A | 4e-23 | 76 | 170 | 4 | 102 | Developmental protein SEPALLATA 3 |
| 4ox0_B | 4e-23 | 76 | 170 | 4 | 102 | Developmental protein SEPALLATA 3 |
| 4ox0_C | 4e-23 | 76 | 170 | 4 | 102 | Developmental protein SEPALLATA 3 |
| 4ox0_D | 4e-23 | 76 | 170 | 4 | 102 | Developmental protein SEPALLATA 3 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in flower development. {ECO:0000250|UniProtKB:Q0HA25}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00315 | DAP | Transfer from AT2G45650 | Download |
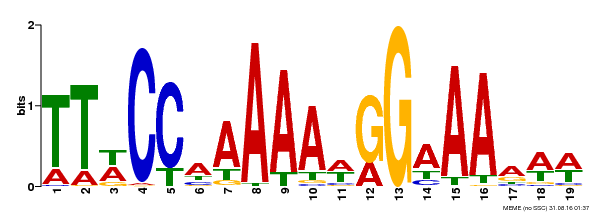
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019259120.1 | 1e-177 | PREDICTED: MADS-box transcription factor 6 | ||||
| Swissprot | Q8LLR1 | 1e-137 | MADS3_VITVI; Agamous-like MADS-box protein MADS3 | ||||
| TrEMBL | A0A314LE78 | 1e-176 | A0A314LE78_NICAT; Mads-box transcription factor 6 | ||||
| STRING | XP_009588019.1 | 1e-175 | (Nicotiana tomentosiformis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA40 | 24 | 625 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G45650.1 | 2e-92 | AGAMOUS-like 6 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



