 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Niben101Scf00698g02020.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Nicotianoideae; Nicotianeae; Nicotiana
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 520aa MW: 57901.8 Da PI: 6.7344 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 54 | 3.1e-17 | 340 | 386 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+ellP++ +K +Ka++L +A+eY+ksLq
Niben101Scf00698g02020.1 340 VHNLSERRRRDRINEKMKALQELLPHS-----TKTDKASMLDEAIEYLKSLQ 386
6*************************9.....9******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 3.93E-20 | 334 | 401 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.476 | 336 | 385 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 2.26E-18 | 339 | 390 | No hit | No description |
| Pfam | PF00010 | 1.3E-14 | 340 | 386 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 4.7E-20 | 340 | 394 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 5.0E-18 | 342 | 391 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009585 | Biological Process | red, far-red light phototransduction | ||||
| GO:0009693 | Biological Process | ethylene biosynthetic process | ||||
| GO:0010244 | Biological Process | response to low fluence blue light stimulus by blue low-fluence system | ||||
| GO:0010600 | Biological Process | regulation of auxin biosynthetic process | ||||
| GO:0010928 | Biological Process | regulation of auxin mediated signaling pathway | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 520 aa Download sequence Send to blast |
MLEFLLCAGW FFGTFNLSEI LCSVVNSSFF QSVVLAMNPY LPEWNTEAEL PVPHQKKPMG 60 FDHELVELLW RNGEVVLHSQ THKKQQGYDP NECRQFNKHD QQTIRDVVSC GNHTNLIQDD 120 ETISWLNCPI DESFEKEFCS PFLSEISTNP IEADQSIRQS EDNKAFKFDP LEIKHVFPHS 180 HHSSFDPNPM PPPRFHNSVS AQKSVKNNLK EGSVMTVGSS HCGRNQVAID ADTSRFSSSA 240 NVGLSAAMIT DYTGKVSPQS DTMDRDTFEP ANTSSSGGSG CSYARTCNLS AATNSQSHKR 300 KSRDGEEPEC QSKAGELESA GGNKLAQKSG TARRSRAAEV HNLSERRRRD RINEKMKALQ 360 ELLPHSTKTD KASMLDEAIE YLKSLQMQLQ MMWMGSGMAS MMFPGVQHYM SRIGMGMGPP 420 TVPSIHNAMH LARLPLVDST IPMNTAAPNQ ATICQNSMLN SVNYQRHLHN PNFQDQYASY 480 MGFHPLQGAS QQMNIFGLGS QTAQQSQQLP HPTNTNAPNS |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 344 | 349 | ERRRRD |
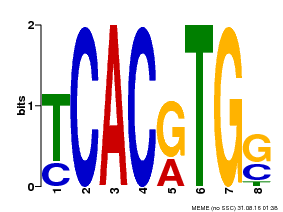
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00082 | ChIP-seq | Transfer from AT3G59060 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009757612.1 | 0.0 | PREDICTED: transcription factor PIF4 | ||||
| Refseq | XP_009757613.1 | 0.0 | PREDICTED: transcription factor PIF4 | ||||
| Refseq | XP_016471316.1 | 0.0 | PREDICTED: transcription factor PIF4-like | ||||
| Refseq | XP_016471375.1 | 0.0 | PREDICTED: transcription factor PIF4-like | ||||
| TrEMBL | A0A1S4A419 | 0.0 | A0A1S4A419_TOBAC; transcription factor PIF4-like | ||||
| TrEMBL | A0A1U7UTT3 | 0.0 | A0A1U7UTT3_NICSY; transcription factor PIF4 | ||||
| STRING | XP_009757612.1 | 0.0 | (Nicotiana sylvestris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA10465 | 22 | 26 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G43010.2 | 1e-48 | phytochrome interacting factor 4 | ||||



