 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Niben101Scf02357g01004.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Nicotianoideae; Nicotianeae; Nicotiana
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 302aa MW: 34710 Da PI: 9.1282 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 98.8 | 2.1e-31 | 69 | 118 | 1 | 50 |
S---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE CS
SRF-TF 1 krienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeys 50
krien++nrqvtf+kRrng+lKKA+ELSvLCdaeva+i+fss+g+lyey+
Niben101Scf02357g01004.1 69 KRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYA 118
79***********************************************8 PP
| |||||||
| 2 | K-box | 108.3 | 8.9e-36 | 137 | 233 | 4 | 100 |
K-box 4 ssgksleeakaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelq 88
+s+ s++ea+++++qqe+akL+++i+ +q+ +R+++Ge L+sLs ++L++Le +Lek++ ++RskKnell+++ie +qk+e e+q
Niben101Scf02357g01004.1 137 TSQGSVSEANTQYYQQEAAKLRRQIRDIQTYNRQIVGEALSSLSPRDLKNLEGKLEKAIGRVRSKKNELLFSEIEVMQKREIEMQ 221
455569******************************************************************************* PP
K-box 89 eenkaLrkklee 100
++n++Lr+k++e
Niben101Scf02357g01004.1 222 NANMYLRAKIAE 233
*********986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00432 | 1.1E-40 | 61 | 120 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS50066 | 33.263 | 61 | 121 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 3.22E-45 | 62 | 138 | No hit | No description |
| SuperFamily | SSF55455 | 3.4E-33 | 62 | 137 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 7.4E-33 | 63 | 83 | IPR002100 | Transcription factor, MADS-box |
| PROSITE pattern | PS00350 | 0 | 63 | 117 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 1.3E-26 | 70 | 117 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 7.4E-33 | 83 | 98 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 7.4E-33 | 98 | 119 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF01486 | 2.2E-25 | 145 | 231 | IPR002487 | Transcription factor, K-box |
| PROSITE profile | PS51297 | 14.515 | 147 | 237 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0048440 | Biological Process | carpel development | ||||
| GO:0048443 | Biological Process | stamen development | ||||
| GO:0048497 | Biological Process | maintenance of floral organ identity | ||||
| GO:0090376 | Biological Process | seed trichome differentiation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 302 aa Download sequence Send to blast |
MEPAKSKEGN AERDEGDHDE DSGKGIAKTY TQKVVLRLDA LERKMEFPNE EFESSNSQRK 60 SGRGKIEIKR IENTTNRQVT FCKRRNGLLK KAYELSVLCD AEVALIVFSS RGRLYEYANN 120 SVRATIDRYK KHHADSTSQG SVSEANTQYY QQEAAKLRRQ IRDIQTYNRQ IVGEALSSLS 180 PRDLKNLEGK LEKAIGRVRS KKNELLFSEI EVMQKREIEM QNANMYLRAK IAEVERAQQQ 240 MNLMPGGSEY SHHQQQPMST SQNYNDARNF LPVNLLEPNP HYSRHDDQTA LQLVGIEIVI 300 LN |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1egw_A | 4e-20 | 62 | 133 | 1 | 72 | MADS BOX TRANSCRIPTION ENHANCER FACTOR 2, POLYPEPTIDE A |
| 1egw_B | 4e-20 | 62 | 133 | 1 | 72 | MADS BOX TRANSCRIPTION ENHANCER FACTOR 2, POLYPEPTIDE A |
| 1egw_C | 4e-20 | 62 | 133 | 1 | 72 | MADS BOX TRANSCRIPTION ENHANCER FACTOR 2, POLYPEPTIDE A |
| 1egw_D | 4e-20 | 62 | 133 | 1 | 72 | MADS BOX TRANSCRIPTION ENHANCER FACTOR 2, POLYPEPTIDE A |
| 3kov_A | 4e-20 | 62 | 145 | 1 | 86 | Myocyte-specific enhancer factor 2A |
| 3kov_B | 4e-20 | 62 | 145 | 1 | 86 | Myocyte-specific enhancer factor 2A |
| 3kov_I | 4e-20 | 62 | 145 | 1 | 86 | Myocyte-specific enhancer factor 2A |
| 3kov_J | 4e-20 | 62 | 145 | 1 | 86 | Myocyte-specific enhancer factor 2A |
| 3p57_A | 4e-20 | 62 | 145 | 1 | 86 | Myocyte-specific enhancer factor 2A |
| 3p57_B | 4e-20 | 62 | 145 | 1 | 86 | Myocyte-specific enhancer factor 2A |
| 3p57_C | 4e-20 | 62 | 145 | 1 | 86 | Myocyte-specific enhancer factor 2A |
| 3p57_D | 4e-20 | 62 | 145 | 1 | 86 | Myocyte-specific enhancer factor 2A |
| 3p57_I | 4e-20 | 62 | 145 | 1 | 86 | Myocyte-specific enhancer factor 2A |
| 3p57_J | 4e-20 | 62 | 145 | 1 | 86 | Myocyte-specific enhancer factor 2A |
| 5f28_A | 4e-20 | 62 | 145 | 2 | 87 | MEF2C |
| 5f28_B | 4e-20 | 62 | 145 | 2 | 87 | MEF2C |
| 5f28_C | 4e-20 | 62 | 145 | 2 | 87 | MEF2C |
| 5f28_D | 4e-20 | 62 | 145 | 2 | 87 | MEF2C |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor involved in flower development. {ECO:0000250|UniProtKB:Q0HA25}. | |||||
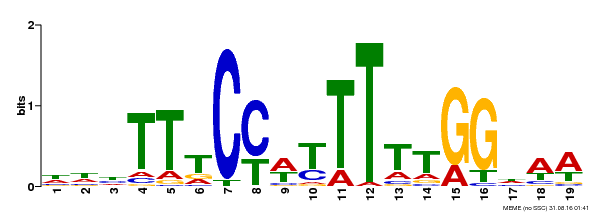
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00609 | ChIP-seq | Transfer from AT4G18960 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009762633.1 | 0.0 | PREDICTED: floral homeotic protein AGAMOUS isoform X2 | ||||
| Refseq | XP_009762634.1 | 0.0 | PREDICTED: floral homeotic protein AGAMOUS isoform X3 | ||||
| Refseq | XP_016486565.1 | 0.0 | PREDICTED: floral homeotic protein AGAMOUS-like isoform X2 | ||||
| Swissprot | Q93XH4 | 1e-124 | MADS1_VITVI; Agamous-like MADS-box protein MADS1 | ||||
| TrEMBL | I3QNW2 | 0.0 | I3QNW2_NICBE; Shatterproof | ||||
| STRING | XP_009762632.1 | 1e-178 | (Nicotiana sylvestris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA40 | 24 | 625 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G18960.1 | 1e-112 | MIKC_MADS family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



