 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Niben101Scf02688g00009.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Nicotianoideae; Nicotianeae; Nicotiana
|
||||||||
| Family | EIL | ||||||||
| Protein Properties | Length: 655aa MW: 73093.6 Da PI: 6.7573 | ||||||||
| Description | EIL family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | EIN3 | 417.9 | 1.1e-127 | 38 | 373 | 1 | 316 |
XXXXXXXXXXXXXXXXXXXXXXX..XXXXX.XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX CS
EIN3 1 eelkkrmwkdqmllkrlkerkkqlledkeaatgakksnksneqarrkkmsraQDgiLkYMlkemevcnaqGfvYgiipekgkpve 85
eel++rmwkd+++lkrlker+k + +a++++++++ ++qarrkkmsraQDgiLkYMlk mevcna+GfvYgiip+kgkpv+
Niben101Scf02688g00009.1 38 EELERRMWKDRIKLKRLKERQKLA---ALKAAEKQNNKHVTDQARRKKMSRAQDGILKYMLKLMEVCNARGFVYGIIPDKGKPVS 119
8*******************9964...34457899999*********************************************** PP
XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX....XX----STTS-HHHHHHHHHHHSSSSSS-TTS--TTT--HH CS
EIN3 86 gasdsLraWWkekvefdrngpaaiskyqaknlilsgesslqtersseshslselqDTtlgSLLsalmqhcdppqrrfplekgvep 170
gasd++raWWkekv+fd+ngpaai+ky+a+ + +++ ++ ++ + + l++lqD+tlgSLLs+lmqhcdppqr++plekgv+p
Niben101Scf02688g00009.1 120 GASDNIRAWWKEKVKFDKNGPAAIAKYEAESRAKGEGVGS--QNGNAGSVLQDLQDATLGSLLSSLMQHCDPPQRKYPLEKGVSP 202
********************************99988888..56888999*********************************** PP
HH---S--HHHHHHT--TT--.-----GGG--HHHHHHHHHHHHHHTGGGHHHHHHTTTTSSSSTTT--SHHHHHHHHHHTTTTT CS
EIN3 171 PWWPtGkelwwgelglskdqgtppykkphdlkkawkvsvLtavikhmsptieeirelerqskylqdkmsakesfallsvlnqeek 255
PWWPtG+e+ww++ gl+k q+ ppykkphdlkk+wkv+vLtavikhmsp+i++ir+l+ qsk+lqdkm+akes ++l+vl++ee+
Niben101Scf02688g00009.1 203 PWWPTGNEEWWSKTGLPKGQN-PPYKKPHDLKKMWKVGVLTAVIKHMSPDIAKIRRLVHQSKCLQDKMTAKESSIWLAVLSKEES 286
*********************.9*************************************************************9 PP
-S--XXXX..XX......XXXXXXXXXXXXXXXXXXXX.........XXXXXX.XXXXXXXXXX....................X CS
EIN3 256 ecatvsah..ss......slrkqspkvtlsceqkedve.........gkkeskikhvqavktta....................g 303
++ ++ ss +r++++k +s+++++dv+ +++++++ g
Niben101Scf02688g00009.1 287 VLQLPGSEngSSsieppaRSRSEKKKPSVSSDSEYDVDgiddgigcvS---------SKDERRNqpldvhplnvipqshqskeqG 362
988777776533678877778899999999********5444433331.........3333333455555555555555444321 PP
XXXXXXXXXXXXX CS
EIN3 304 fpvvrkrkkkpse 316
r rk+k+++
Niben101Scf02688g00009.1 363 --DGRHRKRKRAR 373
..12222222222 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04873 | 7.3E-124 | 38 | 285 | No hit | No description |
| Gene3D | G3DSA:1.10.3180.10 | 5.1E-70 | 163 | 288 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| SuperFamily | SSF116768 | 5.75E-61 | 167 | 287 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042762 | Biological Process | regulation of sulfur metabolic process | ||||
| GO:0071281 | Biological Process | cellular response to iron ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 655 aa Download sequence Send to blast |
MAVIDEIGVD VSSDIEVDDI RCDNIAEKDV SDEEIEPEEL ERRMWKDRIK LKRLKERQKL 60 AALKAAEKQN NKHVTDQARR KKMSRAQDGI LKYMLKLMEV CNARGFVYGI IPDKGKPVSG 120 ASDNIRAWWK EKVKFDKNGP AAIAKYEAES RAKGEGVGSQ NGNAGSVLQD LQDATLGSLL 180 SSLMQHCDPP QRKYPLEKGV SPPWWPTGNE EWWSKTGLPK GQNPPYKKPH DLKKMWKVGV 240 LTAVIKHMSP DIAKIRRLVH QSKCLQDKMT AKESSIWLAV LSKEESVLQL PGSENGSSSI 300 EPPARSRSEK KKPSVSSDSE YDVDGIDDGI GCVSSKDERR NQPLDVHPLN VIPQSHQSKE 360 QGDGRHRKRK RARSNPTEQQ ILPSLRHGDD HSNSSPDTNS SESLRENDKS GAPKPVENSI 420 LTQSELPLQD SNLSLVMSAN VVSIEDAYIG TGPSLYPLHQ NSAVVPYESG VHLESQDSAV 480 RHQFHDAQLH GHHKIPGIKY GPRNSHLHYG SHSSVVHTEL QVSGFAHGSQ ISNFNQPPLY 540 HSYSSAEFEF THEKPQSQMA CNERPIRPRD SGVGSLLHGS GNNISVDNHQ YEKDMSQNNH 600 DRHIEMPFPS PLTIGSPDYA TLGSPFDLGL DVQSYLDNTD FDLDFDKEMM SFFAS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wij_A | 1e-71 | 167 | 296 | 11 | 140 | ETHYLENE-INSENSITIVE3-like 3 protein |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in the ethylene response pathway. {ECO:0000269|PubMed:9215635, ECO:0000269|PubMed:9851977}. | |||||
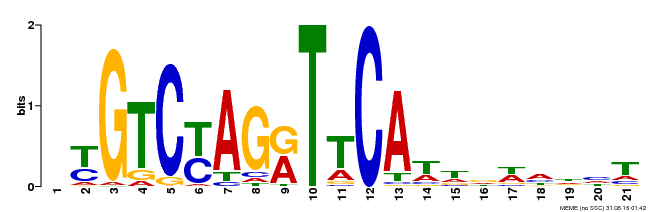
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00233 | DAP | Transfer from AT1G73730 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016477203.1 | 0.0 | PREDICTED: ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Swissprot | O23116 | 1e-169 | EIL3_ARATH; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| TrEMBL | A0A1S4AL79 | 0.0 | A0A1S4AL79_TOBAC; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| STRING | PGSC0003DMT400055102 | 0.0 | (Solanum tuberosum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA304 | 23 | 178 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73730.1 | 1e-149 | ETHYLENE-INSENSITIVE3-like 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



