 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Niben101Scf02807g11013.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Nicotianoideae; Nicotianeae; Nicotiana
|
||||||||
| Family | FAR1 | ||||||||
| Protein Properties | Length: 963aa MW: 109475 Da PI: 6.6765 | ||||||||
| Description | FAR1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | FAR1 | 85 | 1.2e-26 | 91 | 193 | 2 | 91 |
FAR1 2 fYneYAkevGFsvrkskskkskrngeitkrtfvCskegkreeekkk.............tekerrtraetrtgCkaklkvkkekd 73
fY+eYA+++GF++ +++s++sk+++e+++++f Cs++g+++e +k+ e++ +ra +t+Cka+++vk++ d
Niben101Scf02807g11013.1 91 FYQEYARSMGFNTAIQNSRRSKTSREFIDAKFACSRYGTKREYEKSanrprsrqgnkqdPENATGRRACAKTDCKASMHVKRRPD 175
9***************************************999999899999888776555556699****************** PP
FAR1 74 gkwevtkleleHnHelap 91
gkw ++++e+eHnHel p
Niben101Scf02807g11013.1 176 GKWIIHRFEKEHNHELLP 193
***************975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03101 | 1.4E-24 | 91 | 193 | IPR004330 | FAR1 DNA binding domain |
| Pfam | PF10551 | 8.2E-29 | 291 | 383 | IPR018289 | MULE transposase domain |
| PROSITE profile | PS50966 | 9.557 | 571 | 607 | IPR007527 | Zinc finger, SWIM-type |
| Pfam | PF04434 | 7.6E-6 | 572 | 605 | IPR007527 | Zinc finger, SWIM-type |
| SMART | SM00575 | 1.1E-6 | 582 | 609 | IPR006564 | Zinc finger, PMZ-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009585 | Biological Process | red, far-red light phototransduction | ||||
| GO:0010218 | Biological Process | response to far red light | ||||
| GO:0042753 | Biological Process | positive regulation of circadian rhythm | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008270 | Molecular Function | zinc ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 963 aa Download sequence Send to blast |
MDIDLRLPSR DHDKEEEEEQ NGIINMLDNE EKMHSDDGMH GMLVVVEEKM HAEDGGDMNT 60 PVANIIEFKE DVNLEPLAGM EFESHGEAYA FYQEYARSMG FNTAIQNSRR SKTSREFIDA 120 KFACSRYGTK REYEKSANRP RSRQGNKQDP ENATGRRACA KTDCKASMHV KRRPDGKWII 180 HRFEKEHNHE LLPAQAVSEQ TRRMYAAMAR KFAEYKNVVG LKSDSKGPFE KGRNSAMDGG 240 DINILLEFFI QMQSLNSNFF YAVDVGEDQR VKNLIWVDAK ARHDYANFSD VVSFDTTYIR 300 NKYKMPLALF VGVNQHYQFM LLGCALVSDE SATTFSWVML TWLKAMGGQA PKTVITDHDQ 360 VLKSVISEAL PSSLHYFCMW HILGKVSETL NHVIKQNEKF MAKFEKCIYR SSTDEEFEKR 420 WRKLVDRFDL REAELIHSLY EDRVKWIPTF MGDAFLAGMS TVQRSESVNS FFDKYVHKKT 480 TVQEFVKQYE TILQDRYEEE AKADSDTWNK QPALKSPSPF EKHVAGLYTH AVFKKFQAEV 540 LGAVACIPKR EQQDETTITF GVKDYEKDQD FIVTLNEVKS EISCVCHLFE FKGYLCRHAL 600 IVLQICGVSS IPLQYILKRW TKDAKSKYSM TDGSEDVQSR VQRYNELCYR AMKLSEEGSL 660 SQESYSFALR ALDDAFGSCV TFNNSNKNIL EAGTSSAPAL LCVEDDNQSR SMSKTNKKKN 720 SFTKKRKVNS EPDVMAVGAA DSLQQMDKLN SRPVTLDGYF GPQQSVQGMV QLNLMAPTRD 780 NYYGNQQTIQ GLGQLNSIAP THDGYYGAQP TMHGLGRMDF FRTPSFPYGI RVNSEPDVMA 840 VGAADSLQQM DKLNSRPVTL DGYFGPQQSV QGMVQLNLMA PTRDNYYGNQ QTIQGLGQLN 900 SIAPTHDGYY GAQPTMHGLG QMDFFRAPSF PYGIREKVNI FQLELQAVVG STANRKDLCS 960 ACL |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
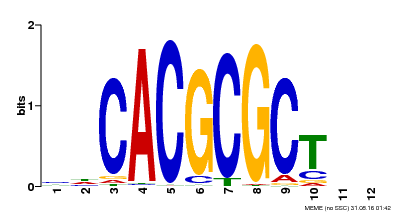
| UniProt | Transcription activator that recognizes and binds to the DNA consensus sequence 5'-CACGCGC-3'. Activates the expression of FHY1 and FHL involved in light responses. When associated with PHYA, protects it from being recognized and degraded by the COP1/SPA complex. Positive regulator of chlorophyll biosynthesis via the activation of HEMB1 gene expression. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:12753585, ECO:0000269|PubMed:17012604, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:18715961, ECO:0000269|PubMed:22634759}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00078 | ChIP-seq | Transfer from AT3G22170 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated after exposure to far-red light. Subject to a negative feedback regulation by PHYA signaling. Up-regulated by white light. {ECO:0000269|PubMed:11889039, ECO:0000269|PubMed:18033885, ECO:0000269|PubMed:22634759}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009804948.1 | 0.0 | PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3 isoform X3 | ||||
| Refseq | XP_009804949.1 | 0.0 | PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3 isoform X3 | ||||
| Refseq | XP_016481998.1 | 0.0 | PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3-like isoform X3 | ||||
| Refseq | XP_016481999.1 | 0.0 | PREDICTED: protein FAR-RED ELONGATED HYPOCOTYL 3-like isoform X3 | ||||
| Swissprot | Q9LIE5 | 0.0 | FHY3_ARATH; Protein FAR-RED ELONGATED HYPOCOTYL 3 | ||||
| TrEMBL | A0A1S4AZK2 | 0.0 | A0A1S4AZK2_TOBAC; protein FAR-RED ELONGATED HYPOCOTYL 3-like isoform X3 | ||||
| TrEMBL | A0A1U7YPD6 | 0.0 | A0A1U7YPD6_NICSY; protein FAR-RED ELONGATED HYPOCOTYL 3 isoform X3 | ||||
| STRING | XP_009804946.1 | 0.0 | (Nicotiana sylvestris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA496 | 21 | 103 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G22170.2 | 0.0 | far-red elongated hypocotyls 3 | ||||



