 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Niben101Scf02862g01006.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Nicotianoideae; Nicotianeae; Nicotiana
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 368aa MW: 42430.4 Da PI: 4.9473 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 176.1 | 9.9e-55 | 16 | 145 | 1 | 129 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLp.k..kvkaeekewyfFskrdkkyatgkrknratksgy 82
+ppGfrFhPtdeelv +yL+kk++++k++l +vi+++d+y++ePwdL+ k ++e++ewyfFs+rdkky+tg+r+nrat +g+
Niben101Scf02862g01006.1 16 VPPGFRFHPTDEELVGYYLRKKIASQKIDL-DVIRDIDLYRIEPWDLRdKcrIGYEEQNEWYFFSHRDKKYPTGTRTNRATVAGF 99
69****************************.9***************843432233667************************** PP
NAM 83 WkatgkdkevlskkgelvglkktLvfykgrapkgektdWvmheyrle 129
Wkatg+dk+v++ k +l+g++ktLvfykgrap+g+k+dW+mheyrle
Niben101Scf02862g01006.1 100 WKATGRDKAVYD-KLKLIGMRKTLVFYKGRAPNGQKSDWIMHEYRLE 145
************.999*****************************85 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 4.18E-60 | 12 | 165 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 58.864 | 16 | 165 | IPR003441 | NAC domain |
| Pfam | PF02365 | 1.7E-28 | 17 | 144 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0048759 | Biological Process | xylem vessel member cell differentiation | ||||
| GO:1901348 | Biological Process | positive regulation of secondary cell wall biogenesis | ||||
| GO:1990110 | Biological Process | callus formation | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 368 aa Download sequence Send to blast |
TKINRSEVMD TNESCVPPGF RFHPTDEELV GYYLRKKIAS QKIDLDVIRD IDLYRIEPWD 60 LRDKCRIGYE EQNEWYFFSH RDKKYPTGTR TNRATVAGFW KATGRDKAVY DKLKLIGMRK 120 TLVFYKGRAP NGQKSDWIMH EYRLESDENG PPQAKGWVVC RAFKKKITGQ TKTNMGEGWE 180 SNYFYEEATT AVSSVVDPLE YITRQPPPPN FMAQNLLCKQ EMELAENNNL NFAYSNSDQF 240 VQLPQLESPT LPLIKRPISS MSLVSENSNQ EDDEQTINRR CSTSQNVTGD WRALDKFVAA 300 QLSHDNHEQE STYEADHGLL SSFEEEARNY SDLGMLLLLS ERTEGPKLNE ILSSTSDDFD 360 TGVCIFDK |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 1e-49 | 16 | 170 | 17 | 170 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 1e-49 | 16 | 170 | 17 | 170 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 1e-49 | 16 | 170 | 17 | 170 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 1e-49 | 16 | 170 | 17 | 170 | NO APICAL MERISTEM PROTEIN |
| 4dul_A | 1e-49 | 16 | 170 | 17 | 170 | NAC domain-containing protein 19 |
| 4dul_B | 1e-49 | 16 | 170 | 17 | 170 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the secondary wall NAC binding element (SNBE), 5'-(T/A)NN(C/T)(T/C/G)TNNNNNNNA(A/C)GN(A/C/T)(A/T)-3', in the promoter of target genes (By similarity). Involved in xylem formation by promoting the expression of secondary wall-associated transcription factors and of genes involved in secondary wall biosynthesis and programmed cell death, genes driven by the secondary wall NAC binding element (SNBE). Triggers thickening of secondary walls (PubMed:25148240). {ECO:0000250|UniProtKB:Q9LVA1, ECO:0000269|PubMed:25148240}. | |||||
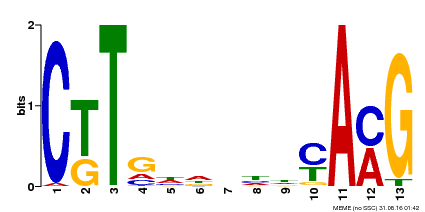
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00266 | DAP | Transfer from AT2G18060 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by chitin (e.g. chitooctaose) (PubMed:17722694). Accumulates in plants exposed to callus induction medium (CIM) (PubMed:17581762). {ECO:0000269|PubMed:17581762, ECO:0000269|PubMed:17722694}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009782335.1 | 0.0 | PREDICTED: NAC domain-containing protein 7-like | ||||
| Refseq | XP_016469037.1 | 0.0 | PREDICTED: NAC domain-containing protein 37-like | ||||
| Swissprot | Q9SL41 | 1e-156 | NAC37_ARATH; NAC domain-containing protein 37 | ||||
| TrEMBL | A0A1S3ZXB2 | 0.0 | A0A1S3ZXB2_TOBAC; NAC domain-containing protein 37-like | ||||
| TrEMBL | A0A1U7X771 | 0.0 | A0A1U7X771_NICSY; NAC domain-containing protein 7-like | ||||
| STRING | XP_009782335.1 | 0.0 | (Nicotiana sylvestris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA134 | 24 | 297 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G18060.1 | 1e-158 | vascular related NAC-domain protein 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



