 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Niben101Scf05551g03011.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Nicotianoideae; Nicotianeae; Nicotiana
|
||||||||
| Family | Dof | ||||||||
| Protein Properties | Length: 324aa MW: 35881.9 Da PI: 6.7806 | ||||||||
| Description | Dof family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-Dof | 126.5 | 8.1e-40 | 56 | 116 | 2 | 62 |
zf-Dof 2 kekalkcprCdstntkfCyynnyslsqPryfCkaCrryWtkGGalrnvPvGggrrknkkss 62
+++++kcprCdst+tkfCyynnyslsqPryfCk+CrryWtkGG+lrn+PvGgg+rknkk+s
Niben101Scf05551g03011.1 56 HDHSVKCPRCDSTHTKFCYYNNYSLSQPRYFCKTCRRYWTKGGTLRNIPVGGGCRKNKKVS 116
57889*****************************************************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD007478 | 1.0E-29 | 50 | 110 | IPR003851 | Zinc finger, Dof-type |
| Pfam | PF02701 | 2.7E-33 | 58 | 114 | IPR003851 | Zinc finger, Dof-type |
| PROSITE profile | PS50884 | 28.994 | 60 | 114 | IPR003851 | Zinc finger, Dof-type |
| PROSITE pattern | PS01361 | 0 | 62 | 98 | IPR003851 | Zinc finger, Dof-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010067 | Biological Process | procambium histogenesis | ||||
| GO:0010087 | Biological Process | phloem or xylem histogenesis | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 324 aa Download sequence Send to blast |
MGITSLQVCM DSSDWLQGTI HEEGAAGIDS SSSPISEDIL TCSRPLIVER RLRPPHDHSV 60 KCPRCDSTHT KFCYYNNYSL SQPRYFCKTC RRYWTKGGTL RNIPVGGGCR KNKKVSSKKS 120 NIETANNQNP TDLQLSFPDN MPPFSHHHFM SSNNFGLGNH QGNNFMLENN TPIDFMESKY 180 EALVGCTSSR NQDFPGNGDI GMISTPTGFG HDINSHGNII APNFAFGMAT MDAGNNFGTT 240 EMNSSHHHQR LMLSNYENNQ HEEQIINAVV KPNPKILSLE WHDQLQGCSD AGKESFSYYS 300 IVGGLGSWTG LVNDYGSSAT NPLV |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence (By similarity). Promotes expression (PubMed:19915089). The PEAR proteins (e.g. DOF2.4, DOF5.1, DOF3.2, DOF1.1, DOF5.6 and DOF5.3) activate gene expression that promotes radial growth of protophloem sieve elements (PubMed:30626969). Involved in the regulation of interfascicular cambium formation and vascular tissue development, particularly at a very early stage during inflorescence stem development; promotes both cambium activity and phloem specification, but prevents xylem specification (PubMed:19915089). {ECO:0000250|UniProtKB:Q9M2U1, ECO:0000269|PubMed:19915089, ECO:0000269|PubMed:30626969}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00580 | DAP | Transfer from AT5G62940 | Download |
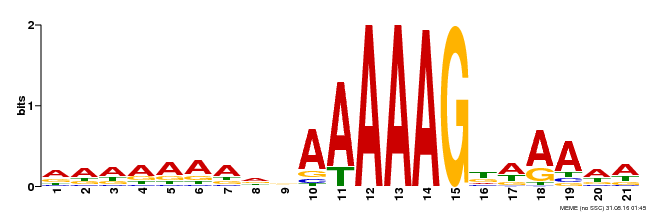
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019240135.1 | 0.0 | PREDICTED: dof zinc finger protein DOF5.6-like | ||||
| Swissprot | Q9FM03 | 4e-49 | DOF56_ARATH; Dof zinc finger protein DOF5.6 | ||||
| TrEMBL | A0A1J6K5Z6 | 0.0 | A0A1J6K5Z6_NICAT; Dof zinc finger protein dof5.6 | ||||
| STRING | XP_009765174.1 | 0.0 | (Nicotiana sylvestris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4895 | 22 | 40 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G62940.1 | 3e-45 | Dof family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



