 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Niben101Scf05552g08002.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Nicotianoideae; Nicotianeae; Nicotiana
|
||||||||
| Family | TCP | ||||||||
| Protein Properties | Length: 727aa MW: 78272.3 Da PI: 5.1419 | ||||||||
| Description | TCP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCP | 117.8 | 1.4e-36 | 430 | 525 | 2 | 97 |
TCP 2 agkkdrhskihTkvggRdRRvRlsaecaarfFdLqdeLGfdkdsktieWLlqqakpaikeltgtssssasec........eaess 78
+++k++++++hTkv+gR+RR+R++a caar+F+L++eLG+++d++ti WLl+ a+pai+e+tgt++ +a ++
Niben101Scf05552g08002.1 430 PPAKRSSKDRHTKVEGRGRRIRMPAACAARIFQLTRELGHKSDGETIRWLLERAEPAIIEATGTGTVPAIAVsvngtlkiP---- 510
6799************************************************************77777444211111110.... PP
TCP 79 sssasnsssgkaaksaaks 97
+ ++ + ++ ++a++
Niben101Scf05552g08002.1 511 ---TTSPATN-SEGENATK 525
...1111111.11111111 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF01730 | 3.9E-19 | 122 | 267 | IPR002639 | Urease accessory protein UreF |
| Pfam | PF03634 | 1.7E-29 | 435 | 516 | IPR005333 | Transcription factor, TCP |
| PROSITE profile | PS51369 | 27.342 | 436 | 490 | IPR017887 | Transcription factor TCP subgroup |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006807 | Biological Process | nitrogen compound metabolic process | ||||
| GO:1905182 | Biological Process | positive regulation of urease activity | ||||
| GO:0016151 | Molecular Function | nickel cation binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 727 aa Download sequence Send to blast |
MDFQFGLGSG PSKMEDVVEK IDDALQFEPN GSLLQWSQWQ LLDSVLPTGG FAHSFGLEAA 60 IQARPSKMED VVEKIDDALQ FEPNGSLLQW SQWQLLDSVL PTGGFAHSFG LEAAIQARLV 120 SGPEDLRTFV THLLENTGSL LLPFIYTLNA SPNVETWYKL DKILDATLTN EVGRKASISQ 180 GSALLRVAAA VFQEVPSLKT MREMSLTTGA VRFHHAPIFG LVCGLLGLNA ETSQKAYMFT 240 TMRDVVSAAT RLNLVGPLGA AVLQHQLAAS AEELSKKWMN RPVQEACQTS PLLDTVQGCH 300 EAPTTAMASL RTAEGYMVDS EWPLDNSKDG KVGLFLAFFF NMEDKQKIEP ENDEVNDSKI 360 LSADPLLGDS EEYTITAVGQ HVTDAEGVGP SQPPSRDIVP LLKEEPAEND LRGGTMSVGL 420 NMQLQKVEKP PAKRSSKDRH TKVEGRGRRI RMPAACAARI FQLTRELGHK SDGETIRWLL 480 ERAEPAIIEA TGTGTVPAIA VSVNGTLKIP TTSPATNSEG ENATKRRKRA ANSEFYDVHE 540 PSNFAPVAPI APQSLLPVWA VASANGLVPT TSGTTFFMVQ PPPTSTTTIA TAGPSYPPQL 600 WATPLYNIPG RPISNYVSGI SYAGVSAAEG QVSSSKSDGN VSAMAPSSSS TGVTTTTTTS 660 TATTQMLRDF SLEIYDKREL QFMHNVGSSC RSGGAKASMG SSPTSWSMGP SSLSKAAAWW 720 STAYDYY |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00667 | PBM | Transfer from GSVIVG01027588001 | Download |
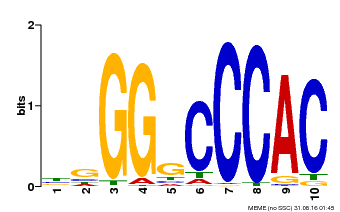
| |||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G51910.2 | 3e-63 | TCP family protein | ||||



