 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Niben101Scf06603g03002.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Nicotianoideae; Nicotianeae; Nicotiana
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 320aa MW: 36208.8 Da PI: 5.4135 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 75.7 | 5.5e-24 | 113 | 174 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-ST...T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsa...gCpvkkkversaedpkvveitYegeHnhe 59
+dDg++WrKYGqK++ s++pr+Y+rCt++ +C+++k+v++ +++p ++ +tY g+H+++
Niben101Scf06603g03002.1 113 VDDGHAWRKYGQKQILHSTYPRHYFRCTHKydqKCQATKQVQKIQDNPPLFRTTYYGNHTCK 174
59***************************99999**************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 1.6E-25 | 98 | 174 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 19.382 | 108 | 171 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 9.02E-24 | 109 | 175 | IPR003657 | WRKY domain |
| SMART | SM00774 | 2.9E-35 | 113 | 175 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 5.9E-23 | 114 | 174 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009862 | Biological Process | systemic acquired resistance, salicylic acid mediated signaling pathway | ||||
| GO:0009864 | Biological Process | induced systemic resistance, jasmonic acid mediated signaling pathway | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:1900056 | Biological Process | negative regulation of leaf senescence | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 320 aa Download sequence Send to blast |
MESPLPEKSS ADLKRAIDGL IRGQEFTRQL KEIIKQPLAT IMAEDLVGKI MNSFSETLSV 60 INSGESDEDT AEVKSPEDSS GSCKSTTSFK DRRGCYKRRK TSETNTKETS DLVDDGHAWR 120 KYGQKQILHS TYPRHYFRCT HKYDQKCQAT KQVQKIQDNP PLFRTTYYGN HTCKPFPRVS 180 QMILDTPIHG DSSILLSFDR NNNNYSSVQN VNHNYQPYNI PTFPSIKQET NEEVFQRSCT 240 SYPKIEDQNQ SSNSDYSLLA NDDHLSTPAA FEASGGHMAA ALSTEVISSG VYSSCTTSTD 300 NLEIDFDFEE SLWNFEGYNS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6ir8_A | 2e-22 | 113 | 174 | 8 | 69 | OsWRKY45 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in senescence, biotic and abiotic stress responses by modulating various phytohormones signaling pathways (By similarity). Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (By similarity). Required for the resistance (R) gene Mi-1-mediated resistance to potato aphid (e.g. M.euphorbiae) and root-knot nematode (e.g. M.javanica) (PubMed:21898085). {ECO:0000250|UniProtKB:Q9LY00, ECO:0000250|UniProtKB:Q9SUP6, ECO:0000269|PubMed:21898085}. | |||||
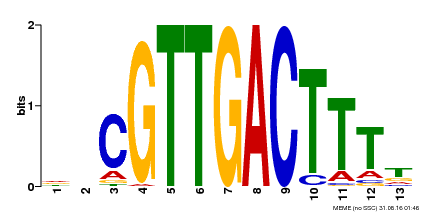
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00408 | DAP | Transfer from AT3G56400 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced in response to potato aphid (e.g. M.euphorbiae) infestation and root-knot nematode (e.g. M.javanica) inoculation. Up-regulated by salicylic acid (SA) and suppressed by methyl jasmonate (MeJA). {ECO:0000269|PubMed:21898085}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009597703.1 | 0.0 | PREDICTED: probable WRKY transcription factor 70 | ||||
| Refseq | XP_016436463.1 | 0.0 | PREDICTED: probable WRKY transcription factor 70 | ||||
| Swissprot | K4BIZ9 | 4e-52 | WRK70_SOLLC; WRKY DNA-binding transcription factor 70 | ||||
| TrEMBL | A0A1S3X9M4 | 0.0 | A0A1S3X9M4_TOBAC; probable WRKY transcription factor 70 | ||||
| STRING | XP_009597703.1 | 0.0 | (Nicotiana tomentosiformis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4230 | 21 | 43 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G56400.1 | 4e-44 | WRKY DNA-binding protein 70 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



