 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Niben101Scf07673g04009.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Nicotianoideae; Nicotianeae; Nicotiana
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 331aa MW: 38237.1 Da PI: 8.8067 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 159.3 | 1.5e-49 | 35 | 170 | 2 | 128 |
NAM 2 ppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpk..kvkaeekewyfFskrdkkyatgkrknratksgyWk 84
pGfrFhPtdeelv++yLk++ve+k++++ + ikevdiyk++PwdLpk +v ++ kewyfFs r +ky+++ r+nr+t sg+Wk
Niben101Scf07673g04009.1 35 LPGFRFHPTDEELVSFYLKRRVENKRIKM-DLIKEVDIYKHDPWDLPKgnTVGGNCKEWYFFSMRGRKYRNSVRPNRVTGSGFWK 118
59***************************.99***************5346666778**************************** PP
NAM 85 atgkdkevlsk..kgel........vglkktLvfykgrapkgektdWvmheyrl 128
atg dk+v+s+ + + +glkk+Lv+y+g+a kg+ktdW+mhe+rl
Niben101Scf07673g04009.1 119 ATGIDKPVFSSttT--TsqctdrdcIGLKKSLVYYRGSAGKGTKTDWMMHEFRL 170
**********8651..344556666***************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51005 | 51.087 | 34 | 203 | IPR003441 | NAC domain |
| SuperFamily | SSF101941 | 2.22E-51 | 34 | 202 | IPR003441 | NAC domain |
| Pfam | PF02365 | 5.0E-25 | 36 | 170 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005992 | Biological Process | trehalose biosynthetic process | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0006561 | Biological Process | proline biosynthetic process | ||||
| GO:0009718 | Biological Process | anthocyanin-containing compound biosynthetic process | ||||
| GO:0010120 | Biological Process | camalexin biosynthetic process | ||||
| GO:0042538 | Biological Process | hyperosmotic salinity response | ||||
| GO:1900056 | Biological Process | negative regulation of leaf senescence | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 331 aa Download sequence Send to blast |
MSEKRAEKIE NMDLVENIST ATNIKAEDEE EIMQLPGFRF HPTDEELVSF YLKRRVENKR 60 IKMDLIKEVD IYKHDPWDLP KGNTVGGNCK EWYFFSMRGR KYRNSVRPNR VTGSGFWKAT 120 GIDKPVFSST TTTSQCTDRD CIGLKKSLVY YRGSAGKGTK TDWMMHEFRL PPNWKTSNQQ 180 LLNPKSVIIP EAEVWTLCRI FKRTSNYKRF TPDWKQPIIK QSFGDASSKA CSLQSETSDN 240 HSININFKKM EFPHKNNNTA IGGNYQVDQR NTDCNNSQRI ITMLQSSITS SNSSFWNTSA 300 EEQYLFSDGN WDELKSVVDL AIDPCSLFGF R |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3ulx_A | 9e-47 | 29 | 209 | 10 | 174 | Stress-induced transcription factor NAC1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds to the 5'- RRYGCCGT-3' consensus core sequence. Central longevity regulator. Negative regulator of leaf senescence. Modulates cellular H(2)O(2) levels and enhances tolerance to various abiotic stresses through the regulation of DREB2A. {ECO:0000269|PubMed:22345491}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00310 | DAP | Transfer from AT2G43000 | Download |
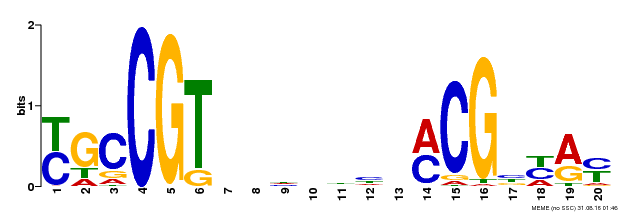
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by H(2)O(2), paraquat, ozone, 3-aminotriazole and salt stress. {ECO:0000269|PubMed:22345491}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009760718.1 | 0.0 | PREDICTED: transcription factor JUNGBRUNNEN 1-like isoform X1 | ||||
| Swissprot | Q9SK55 | 2e-88 | NAC42_ARATH; Transcription factor JUNGBRUNNEN 1 | ||||
| TrEMBL | A0A1U7V2V3 | 0.0 | A0A1U7V2V3_NICSY; transcription factor JUNGBRUNNEN 1-like isoform X1 | ||||
| STRING | XP_009760718.1 | 0.0 | (Nicotiana sylvestris) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA1697 | 24 | 69 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G43000.1 | 5e-87 | NAC domain containing protein 42 | ||||



