 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Niben101Scf09075g02003.1 | ||||||||
| Common Name | HB1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Nicotianoideae; Nicotianeae; Nicotiana
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 243aa MW: 27377.8 Da PI: 9.2273 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 56.9 | 3.7e-18 | 99 | 153 | 2 | 56 |
T--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 2 rkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
rk+ ++tk q Lee F+++ +++ ++++eLA++l+L rqV vWFqNrRa+ k
Niben101Scf09075g02003.1 99 RKKFRLTKAQSALLEESFKQHTTLNPKQKQELARNLNLRPRQVEVWFQNRRARTK 153
678899***********************************************98 PP
| |||||||
| 2 | HD-ZIP_I/II | 122.8 | 1.7e-39 | 99 | 187 | 1 | 90 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeve 85
+kk rl+k q++lLEesF+++++L+p++K+elar+L+l+prqv+vWFqnrRARtk+kq+E+d+e Lk+++++l+een+rL+ke +
Niben101Scf09075g02003.1 99 RKKFRLTKAQSALLEESFKQHTTLNPKQKQELARNLNLRPRQVEVWFQNRRARTKLKQTEVDCEILKKCCETLTEENRRLHKELQ 183
699********************************************************************************** PP
HD-ZIP_I/II 86 eLree 90
eL+ +
Niben101Scf09075g02003.1 184 ELK-A 187
**9.4 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 6.27E-19 | 86 | 156 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 5.3E-18 | 92 | 158 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.605 | 95 | 155 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 2.0E-15 | 97 | 159 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 1.2E-15 | 99 | 153 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 2.33E-15 | 99 | 156 | No hit | No description |
| PROSITE pattern | PS00027 | 0 | 130 | 153 | IPR017970 | Homeobox, conserved site |
| SMART | SM00340 | 8.6E-23 | 155 | 198 | IPR003106 | Leucine zipper, homeobox-associated |
| Pfam | PF02183 | 2.3E-11 | 155 | 188 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009735 | Biological Process | response to cytokinin | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 243 aa Download sequence Send to blast |
MADQKSTKIA PVVMAKGPSL GFEPSLTLSL SGDHTYNKQQ AVKNDHQSAD LYRQDSAASS 60 YSNASVKRER DVGSEEATTE VERVSSRVSD EDDDGSNARK KFRLTKAQSA LLEESFKQHT 120 TLNPKQKQEL ARNLNLRPRQ VEVWFQNRRA RTKLKQTEVD CEILKKCCET LTEENRRLHK 180 ELQELKAVKI AQPLYMQRPA ATLTMCPSCE RIGGVGENTS KNPFTLAQKP HFYNSFTNPS 240 AAC |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 147 | 155 | RRARTKLKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00477 | DAP | Transfer from AT4G37790 | Download |
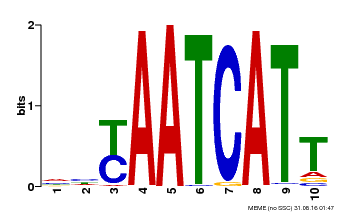
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009592889.1 | 1e-157 | PREDICTED: homeobox-leucine zipper protein HAT22-like | ||||
| Refseq | XP_016496679.1 | 1e-157 | PREDICTED: homeobox-leucine zipper protein HAT22-like | ||||
| Swissprot | P46604 | 2e-87 | HAT22_ARATH; Homeobox-leucine zipper protein HAT22 | ||||
| TrEMBL | C6FFS4 | 0.0 | C6FFS4_NICBE; Homeodomain leucine-zipper 1 | ||||
| STRING | XP_009592889.1 | 1e-157 | (Nicotiana tomentosiformis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA290 | 24 | 186 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G37790.1 | 1e-83 | HD-ZIP family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



