 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Niben101Scf12852g03016.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Nicotianoideae; Nicotianeae; Nicotiana
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 716aa MW: 76711.9 Da PI: 6.9137 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 50 | 5.5e-16 | 443 | 489 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K +Ka++L +A+eY+k Lq
Niben101Scf12852g03016.1 443 VHNLSERRRRDRINEKMRALQELIPNC-----NKADKASMLDEAIEYLKTLQ 489
6*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 4.46E-17 | 435 | 493 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 2.9E-20 | 436 | 497 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 2.09E-20 | 436 | 499 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.228 | 439 | 488 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 2.4E-13 | 443 | 489 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 2.1E-17 | 445 | 494 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 716 aa Download sequence Send to blast |
MPLSEFLKMA RGKFESGQQK VSSTTHLSSF PENDLVELVW ENGQIMMQGQ SSSAKKSPIP 60 NKLPSNASKI GKFGVMDSML NDMPLSVPTC ELDLIQEDEG VPWLGYAADD SLQQDYCSQF 120 LPEMSGVTAN EPSGQSEFGL INKGGSSDKM IRDSHSAPFH NAVNFEQRKA SPSSKFSQLC 180 SLAPQKGHPE SGVSDIFSSK VSNTPVSVYG DSSQNQAAVG EVKSIKIQKQ NMPGAGSCSS 240 LLNFSHFSRP ATFVKAAKPQ NIIAGPNVSD SYTVEAKGSK EGENVSKSHN NVNAAAVEDY 300 LSSKKETDLH YPTNVVTYQL ESRASGASFH DRSCRVEQSD NVFRDRSSNS DNTPGQCTGA 360 KAKPDTADGE RNAEHGVVCS SVCSGSSAER GSSDQPHKLK RKSRDNEDSG CPSEDVEEES 420 VGIKKICPTR GGTGSKRSRA AEVHNLSERR RRDRINEKMR ALQELIPNCN KADKASMLDE 480 AIEYLKTLQL QVQIMSMGAG FCVPPMMFPA GVQHMHAAQM PHFSPMGLGM GMGMGMGYGM 540 GMLEMNGRSS GCPIFPIPSV HGSHFPSPPI PASAAYPGVA VSNRHAFAHP GQGLPMPIPR 600 PSLAPLAGQS SAGAAVPSNV ARVGVSVDRP SVPPILDSKN PVHKKNSQIV NNAESSLPIN 660 QTCSQVQATN EILDKSALVQ KNDQLLDVTD SEANNLTSQS NVRGSEAGEF VLEKRL |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 447 | 452 | ERRRRD |
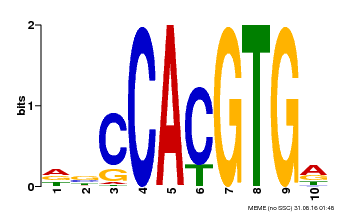
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019258592.1 | 0.0 | PREDICTED: transcription factor PIF3-like isoform X1 | ||||
| TrEMBL | A0A314LFF6 | 0.0 | A0A314LFF6_NICAT; Transcription factor pif3 | ||||
| STRING | XP_009589214.1 | 0.0 | (Nicotiana tomentosiformis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4965 | 24 | 37 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 4e-48 | phytochrome interacting factor 3 | ||||



