 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OB01G34880.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 819aa MW: 93190.6 Da PI: 7.4509 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 104.1 | 7.4e-33 | 167 | 224 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
+Dgy+WrKYGqK vk+s++prsYYrCt+++Cpvkk+vers +dp vv++tYeg+H+h+
OB01G34880.1 167 EDGYRWRKYGQKAVKNSPYPRSYYRCTTQKCPVKKRVERSYQDPAVVITTYEGKHTHP 224
8********************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 1.0E-34 | 151 | 226 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 3.27E-29 | 158 | 226 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 32.535 | 161 | 226 | IPR003657 | WRKY domain |
| SMART | SM00774 | 2.6E-39 | 166 | 225 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 5.5E-26 | 167 | 224 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 819 aa Download sequence Send to blast |
MSGEFQFQDE LAPLFGRQGT TAEMQMPASW FSDYLQAGAP MQMDYDLMCR ALELPVGEDV 60 KREVDVGAGG GGGGVGVVPL TPNTTSSMST SSSEGGGGGG AGEEDSPARC KKEEEEENKE 120 EGKGEEEEGH KNKKGSAAKG GKAGKGEKRQ RQPRFAFMTK SEVDHLEDGY RWRKYGQKAV 180 KNSPYPRSYY RCTTQKCPVK KRVERSYQDP AVVITTYEGK HTHPIPATLR GSTHLLAAHA 240 QAAAHLHHHH GGXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX 300 XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX 360 XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX 420 XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX 480 XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX 540 XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX 600 XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX 660 XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX 720 XXXXXXXXXX XXXXXXXXXX XXXXKKTTMG GAAAAATTHV LTGAIGGGGG VSGATTNAVA 780 VAASSPSPPS LQMQHFMAQD FGLLQDMLLP SFVHGTNQP |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 4e-27 | 157 | 227 | 8 | 78 | Probable WRKY transcription factor 4 |
| 2lex_A | 4e-27 | 157 | 227 | 8 | 78 | Probable WRKY transcription factor 4 |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00440 | DAP | Transfer from AT4G18170 | Download |
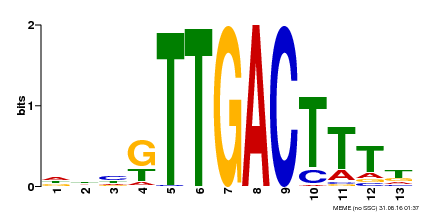
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | OB01G34880.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY341844 | 0.0 | AY341844.1 Oryza sativa (japonica cultivar-group) WRKY3 mRNA, complete cds. | |||
| GenBank | AY341855 | 0.0 | AY341855.1 Oryza sativa (japonica cultivar-group) WRKY14 mRNA, complete cds. | |||
| GenBank | AY870604 | 0.0 | AY870604.1 Oryza sativa (japonica cultivar-group) WRKY16 mRNA, complete cds. | |||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP623 | 37 | 173 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G29860.1 | 3e-46 | WRKY DNA-binding protein 71 | ||||



