 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OB01G48740.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | LBD | ||||||||
| Protein Properties | Length: 273aa MW: 28243.8 Da PI: 8.6578 | ||||||||
| Description | LBD family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF260 | 136.9 | 7.2e-43 | 40 | 138 | 1 | 99 |
DUF260 1 aCaaCkvlrrkCakdCvlapyfpaeqpkkfanvhklFGasnvlkllkalpeeeredamsslvyeAearardPvyGavgvilklqqqleqlkaelall 97
+CaaCk+lrrkC++dCv+apyfp ++p+kf vh++FGasnv+kll++l++ +reda++sl+yeA++r+rdPvyG+v++i+ lq++l+ql+++la++
OB01G48740.1 40 PCAACKFLRRKCQPDCVFAPYFPPDNPQKFVHVHRVFGASNVTKLLNELHPYQREDAVNSLAYEADMRLRDPVYGCVAIISILQRNLRQLQQDLARA 136
7*********************************************************************************************998 PP
DUF260 98 ke 99
k
OB01G48740.1 137 KY 138
76 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50891 | 26.398 | 39 | 140 | IPR004883 | Lateral organ boundaries, LOB |
| Pfam | PF03195 | 1.2E-42 | 40 | 136 | IPR004883 | Lateral organ boundaries, LOB |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009799 | Biological Process | specification of symmetry | ||||
| GO:0009944 | Biological Process | polarity specification of adaxial/abaxial axis | ||||
| GO:0009954 | Biological Process | proximal/distal pattern formation | ||||
| GO:0048441 | Biological Process | petal development | ||||
| GO:0005654 | Cellular Component | nucleoplasm | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 273 aa Download sequence Send to blast |
MASSSASSVP APSGSVITIA SASAAAAAAA NAATCGTGSP CAACKFLRRK CQPDCVFAPY 60 FPPDNPQKFV HVHRVFGASN VTKLLNELHP YQREDAVNSL AYEADMRLRD PVYGCVAIIS 120 ILQRNLRQLQ QDLARAKYEL SKYQAAAAAA AAAAASPPGR RXXXXXXXXX XXXXXXXXXX 180 XXXXXSAALA SVGGGAAACF GPDQSFSAVQ MLSRSYEGEP MARFGGGNGG YEFGYSTSMA 240 GAGHMSSGLG TLGGGPFLKS GIAGSDERPG AGQ |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5ly0_A | 2e-53 | 34 | 143 | 5 | 114 | LOB family transfactor Ramosa2.1 |
| 5ly0_B | 2e-53 | 34 | 143 | 5 | 114 | LOB family transfactor Ramosa2.1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Negative regulator of cell proliferation in the adaxial side of leaves. Regulates the formation of a symmetric lamina and the establishment of venation (By similarity). {ECO:0000250}. | |||||
| UniProt | Negative regulator of cell proliferation in the adaxial side of leaves. Regulates the formation of a symmetric lamina and the establishment of venation (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00211 | DAP | Transfer from AT1G65620 | Download |
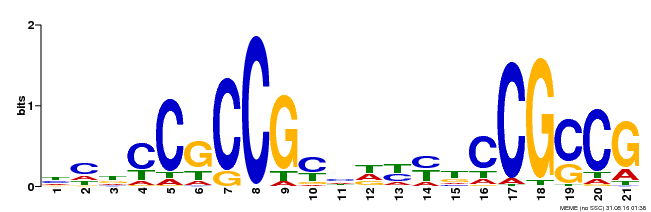
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | OB01G48740.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AB200237 | 0.0 | AB200237.1 Oryza sativa Japonica Group CRLL3 mRNA for LOB domain protein, complete cds. | |||
| GenBank | AK071407 | 0.0 | AK071407.1 Oryza sativa Japonica Group cDNA clone:J023089G14, full insert sequence. | |||
| GenBank | AK119575 | 0.0 | AK119575.1 Oryza sativa Japonica Group cDNA clone:002-117-B04, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006646551.1 | 1e-126 | PREDICTED: LOB domain-containing protein 6-like | ||||
| Swissprot | A2WXT0 | 1e-125 | LBD6_ORYSI; LOB domain-containing protein 6 | ||||
| Swissprot | Q8LQH4 | 1e-125 | LBD6_ORYSJ; LOB domain-containing protein 6 | ||||
| TrEMBL | J3L6L8 | 1e-174 | J3L6L8_ORYBR; Uncharacterized protein | ||||
| STRING | OB01G48740.1 | 1e-175 | (Oryza brachyantha) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2267 | 38 | 95 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G65620.4 | 1e-55 | LBD family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



