 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OB01G50120.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | B3 | ||||||||
| Protein Properties | Length: 731aa MW: 76694.9 Da PI: 7.3278 | ||||||||
| Description | B3 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 59 | 8.3e-19 | 529 | 625 | 2 | 95 |
EEE-..-HHHHTT-EE--HHH.HTT..---..--SEEEEEE.TTS-EEEEEE....EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SS.SEE.. CS
B3 2 fkvltpsdvlksgrlvlpkkfaeeh..ggkkeesktltled.esgrsWevkliy..rkksgryvltkGWkeFvkangLkegDfvvFkldgr.sefel 92
+kvl++sdv+++gr+vlpkk ae h ++k++ ++ + +ed ++++W++++++ ++ks++y+l+ ++ +Fv++n+L+egDf+v++ ++ +++++
OB01G50120.1 529 QKVLKQSDVGSLGRIVLPKKEAEVHlpELKTRDGISIPMEDiGTSQVWNMRYRFwpNNKSRMYLLE-NTGDFVRSNELQEGDFIVIY--SDmKSGKY 622
79*************************999***********7777*******99777777777777.********************..55588877 PP
EEE CS
B3 93 vvk 95
+++
OB01G50120.1 623 LIR 625
765 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.40.330.10 | 1.2E-27 | 522 | 637 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10015 | 9.09E-32 | 526 | 627 | No hit | No description |
| SuperFamily | SSF101936 | 7.72E-22 | 527 | 621 | IPR015300 | DNA-binding pseudobarrel domain |
| SMART | SM01019 | 1.1E-16 | 528 | 630 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 10.292 | 528 | 630 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 2.7E-15 | 529 | 624 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009657 | Biological Process | plastid organization | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009793 | Biological Process | embryo development ending in seed dormancy | ||||
| GO:0031930 | Biological Process | mitochondria-nucleus signaling pathway | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005829 | Cellular Component | cytosol | ||||
| GO:0001076 | Molecular Function | transcription factor activity, RNA polymerase II transcription factor binding | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 731 aa Download sequence Send to blast |
MDASAGSSAP HSQGNAGRQG GGGGRGKAPA AEIRGAEARD DFLFADDTFP SLPDFPCLSS 60 PTSSSFSSSS SSNSSSAFTT AAGGGGGGDG FDELADIDQL LDLASLSSVP WEAEPLFPDD 120 VGMMIEDAIS GQPHQVDCTG EAKAMLEAAG AEDAGGDACM GIEAADDLPA FFMEWLTSNR 180 EYISADDLRS IRLRRSTIEA AAARLGGGRQ GTMQLLKLIL TWVQNHHLQK KRPRTAMDDA 240 AXXGTGQLPS PGANPGYEFP SGGGQEMGSA AATSWMPYQA FTPPAAYGGE AMYPGGAGPY 300 PFQQSCSTSS VVVNSQPFSP PAAAAPAADM HASSGGNMAW PQQFAPFPGS STGSYTMPSV 360 VPQPFTTGFV GQYSGGHAMC SQRLAGVEPS ATKEARKKRM ARQRRLSCLQ QQRSQQINLS 420 QIQISGHPQE PSPRAAHSAP VTPSSGGCRS WPGIWPPAGQ VIQNPLSNKP NNNPSTSKQQ 480 KPSPEKPKPA VSQPAAAVGA QQESPQRSAA SEKRQAATKT DKNLRFLLQK VLKQSDVGSL 540 GRIVLPKKEA EVHLPELKTR DGISIPMEDI GTSQVWNMRY RFWPNNKSRM YLLENTGDFV 600 RSNELQEGDF IVIYSDMKSG KYLIRGVKVR RATQLEQGNN NSSGAVGKHK HGSPEKPGVS 660 NTKAAGAVPG GDDDNDDSVD TAAAGKPDGG CKGGGKSPHG GARRSRQEAA AAASMSQMAP 720 AMADALKLLI I |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j9b_A | 3e-38 | 524 | 637 | 1 | 111 | B3 domain-containing transcription factor FUS3 |
| 6j9b_D | 3e-38 | 524 | 637 | 1 | 111 | B3 domain-containing transcription factor FUS3 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may participate in abscisic acid-regulated gene expression during seed development. May be required for seed maturation and dormancy induction. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00083 | PBM | Transfer from AT3G24650 | Download |
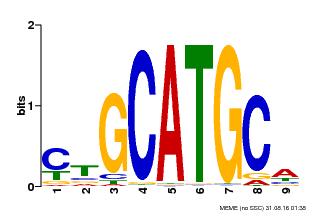
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | OB01G50120.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK073805 | 0.0 | AK073805.1 Oryza sativa Japonica Group cDNA clone:J033069I02, full insert sequence. | |||
| GenBank | AK105441 | 0.0 | AK105441.1 Oryza sativa Japonica Group cDNA clone:001-125-A06, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006645206.1 | 0.0 | PREDICTED: LOW QUALITY PROTEIN: B3 domain-containing protein VP1 | ||||
| Swissprot | P37398 | 0.0 | VIV_ORYSJ; B3 domain-containing protein VP1 | ||||
| TrEMBL | J3L706 | 0.0 | J3L706_ORYBR; Uncharacterized protein | ||||
| STRING | OB01G50120.1 | 0.0 | (Oryza brachyantha) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP7529 | 34 | 43 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G24650.1 | 3e-61 | B3 family protein | ||||



