 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OB02G35920.1 | ||||||||
| Common Name | LOC102700119 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 354aa MW: 37566.8 Da PI: 4.5336 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 52.9 | 8.7e-17 | 16 | 63 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg+WT++Ed +lv +++++G +W++ ++ g+ R++k+c++rw +yl
OB02G35920.1 16 RGSWTPQEDMRLVAYIQKHGHANWRALPKQAGLLRCGKSCRLRWINYL 63
89******************************99************97 PP
| |||||||
| 2 | Myb_DNA-binding | 53.1 | 7.1e-17 | 69 | 114 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg++T+eE+e +++++ +lG++ W++Ia++++ gRt++++k+ w+++l
OB02G35920.1 69 RGNFTAEEEETIIKLHGLLGNK-WSKIASCLP-GRTDNEIKNVWNTHL 114
89********************.*********.************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 14.196 | 11 | 63 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 5.24E-30 | 13 | 110 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.9E-12 | 15 | 65 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 4.8E-15 | 16 | 63 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.2E-23 | 17 | 70 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 4.65E-10 | 18 | 63 | No hit | No description |
| PROSITE profile | PS51294 | 24.612 | 64 | 118 | IPR017930 | Myb domain |
| SMART | SM00717 | 6.3E-15 | 68 | 116 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 5.9E-15 | 69 | 114 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.48E-11 | 71 | 114 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 4.8E-26 | 71 | 117 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:2000652 | Biological Process | regulation of secondary cell wall biogenesis | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 354 aa Download sequence Send to blast |
MGRGRAPCCA KVGLNRGSWT PQEDMRLVAY IQKHGHANWR ALPKQAGLLR CGKSCRLRWI 60 NYLRPDLKRG NFTAEEEETI IKLHGLLGNK WSKIASCLPG RTDNEIKNVW NTHLKKRASQ 120 REQQQQKPGA TKSKDKAVDT SDADTHSPSS SASSSSTTTA NNNGSSSGEQ CGTSREPETV 180 DVSLLEPEID IADMLLVDDA PTEALLAAPM PPSPCSSSSL TTTACIGAVS DDELLDLPEI 240 DIEPEIWSII DGYGGAPVAA GAGDATVPCT TTANAASASP GEEGAEWWVE NLEKELGLWG 300 PMDESLAHPG PLGQVCYAGP LTELTDGDPV STYFQSGPTA SPLPEIASSS AVLS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1mse_C | 2e-26 | 14 | 117 | 2 | 104 | C-Myb DNA-Binding Domain |
| 1msf_C | 2e-26 | 14 | 117 | 2 | 104 | C-Myb DNA-Binding Domain |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that regulates positively genes involved in anthocyanin biosynthesis such as A1. {ECO:0000269|PubMed:7920701}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00249 | DAP | Transfer from AT1G79180 | Download |
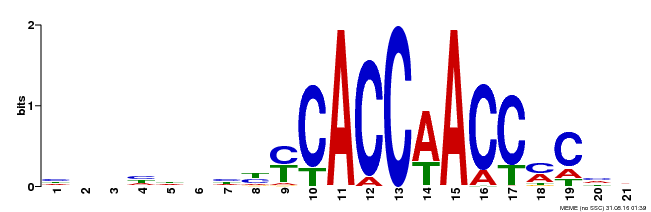
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | OB02G35920.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK111720 | 0.0 | AK111720.1 Oryza sativa Japonica Group cDNA clone:J023030I19, full insert sequence. | |||
| GenBank | AK111933 | 0.0 | AK111933.1 Oryza sativa Japonica Group cDNA clone:001-017-B10, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006647724.1 | 0.0 | PREDICTED: myb-related protein Zm1-like | ||||
| Swissprot | P20024 | 1e-138 | MYB1_MAIZE; Myb-related protein Zm1 | ||||
| TrEMBL | J3LG25 | 0.0 | J3LG25_ORYBR; Uncharacterized protein | ||||
| STRING | OB02G35920.1 | 0.0 | (Oryza brachyantha) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2154 | 37 | 98 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G79180.1 | 3e-69 | myb domain protein 63 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 102700119 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



