 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OB03G25560.1 | ||||||||
| Common Name | LOC102714816 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 355aa MW: 39641 Da PI: 8.0629 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 109.7 | 1.5e-34 | 47 | 101 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
kprl+WtpeLHerFveav+qLGG+ekAtPkti++lm+v+gLtl+h+kSHLQkYRl
OB03G25560.1 47 KPRLKWTPELHERFVEAVNQLGGPEKATPKTIMRLMGVPGLTLYHLKSHLQKYRL 101
79****************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 12.874 | 44 | 104 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 6.5E-33 | 45 | 102 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 6.45E-18 | 46 | 102 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.9E-24 | 47 | 102 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 9.1E-11 | 49 | 100 | IPR001005 | SANT/Myb domain |
| Pfam | PF14379 | 3.3E-25 | 149 | 195 | IPR025756 | MYB-CC type transcription factor, LHEQLE-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 355 aa Download sequence Send to blast |
MYHQQQQIQN HSQLLSSRQS FPSERHLLMQ GGSVPGESGL VLSTDAKPRL KWTPELHERF 60 VEAVNQLGGP EKATPKTIMR LMGVPGLTLY HLKSHLQKYR LSKNLHAQAN VGNVKNALVC 120 TTATEKPSEG NRSPVSHLTL GTQTNKSVHI GEALQMQIEV QRRLHEQLEV QRHLQLRIEA 180 QGKYLQSVLE KAQETLAKQN AGSVGLETAK MELSELVSKV STECLQHAFS GFEEIESSQM 240 LQGHTMHLGD GSVDSCLTVC DGSQKDQDIL SISLSAQKGK EIGCMSFDMH VKERGSEDLF 300 LNKLSRRPSN HQERCERRDG FSMSCQATKL DLNMNDTYGG PKHCKKFDLN GFSWA |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 3e-21 | 46 | 103 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 3e-21 | 46 | 103 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 3e-21 | 47 | 103 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 3e-21 | 47 | 103 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 3e-21 | 47 | 103 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 3e-21 | 47 | 103 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 3e-21 | 46 | 103 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 3e-21 | 46 | 103 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 3e-21 | 46 | 103 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 3e-21 | 46 | 103 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 3e-21 | 46 | 103 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 3e-21 | 46 | 103 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that may activate the transcription of specific genes involved in nitrogen uptake or assimilation (PubMed:15592750). Acts redundantly with MYR1 as a repressor of flowering and organ elongation under decreased light intensity (PubMed:21255164). Represses gibberellic acid (GA)-dependent responses and affects levels of bioactive GA (PubMed:21255164). {ECO:0000269|PubMed:21255164, ECO:0000305|PubMed:15592750}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00331 | DAP | Transfer from AT3G04030 | Download |
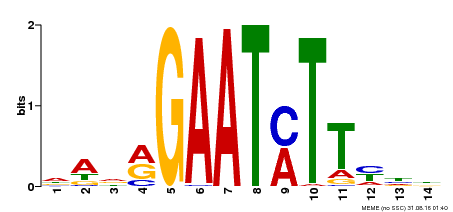
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | OB03G25560.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by nitrogen deficiency. {ECO:0000269|PubMed:15592750}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK100282 | 0.0 | AK100282.1 Oryza sativa Japonica Group cDNA clone:J023074M06, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006650017.1 | 0.0 | PREDICTED: myb family transcription factor APL-like | ||||
| Swissprot | Q9SQQ9 | 1e-106 | PHL9_ARATH; Myb-related protein 2 | ||||
| TrEMBL | J3LND0 | 0.0 | J3LND0_ORYBR; Uncharacterized protein | ||||
| STRING | OB03G25560.1 | 0.0 | (Oryza brachyantha) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3574 | 38 | 79 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G04030.3 | 1e-105 | G2-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 102714816 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



