 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OB04G35540.1 | ||||||||
| Common Name | LOC102720794 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | WOX | ||||||||
| Protein Properties | Length: 287aa MW: 30201.4 Da PI: 8.2036 | ||||||||
| Description | WOX family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 58.2 | 1.4e-18 | 41 | 101 | 3 | 57 |
--SS--HHHHHHHHHHHHH..SSS--HHHHHHHHHHC....TS-HHHHHHHHHHHHHHHHC CS
Homeobox 3 kRttftkeqleeLeelFek..nrypsaeereeLAkkl....gLterqVkvWFqNrRakekk 57
+R+t+t+eq+++L el+++ r p+ e+++++A l +++ ++V++WFqN++a+e++
OB04G35540.1 41 TRWTPTTEQIKILRELYYScgIRSPNSEQIQRIAGMLrqygRIEGKNVFYWFQNHKARERQ 101
7*****************7668*************************************97 PP
| |||||||
| 2 | Wus_type_Homeobox | 112.9 | 1.6e-36 | 40 | 103 | 3 | 65 |
Wus_type_Homeobox 3 rtRWtPtpeQikiLeelyk.sGlrtPnkeeiqritaeLeeyGkiedkNVfyWFQNrkaRerqkq 65
tRWtPt+eQikiL+ely+ +G+r+Pn+e+iqri+ +L++yG+ie+kNVfyWFQN+kaRerqk+
OB04G35540.1 40 GTRWTPTTEQIKILRELYYsCGIRSPNSEQIQRIAGMLRQYGRIEGKNVFYWFQNHKARERQKK 103
69***************9978*****************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 6.42E-12 | 35 | 105 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 10.203 | 36 | 102 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 3.5E-5 | 38 | 106 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 3.7E-16 | 41 | 101 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 2.45E-6 | 41 | 103 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 4.5E-7 | 42 | 101 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010492 | Biological Process | maintenance of shoot apical meristem identity | ||||
| GO:0080166 | Biological Process | stomium development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 287 aa Download sequence Send to blast |
MDRVQPQQQQ QQRQVGGGEE VAGRSGGGGG GGVSVCRPSG TRWTPTTEQI KILRELYYSC 60 GIRSPNSEQI QRIAGMLRQY GRIEGKNVFY WFQNHKARER QKKRLTTLDV NTAAADASQL 120 AFLSLSPTGA TAMAPSFPGP YVGNGGAVSA AQTDQATVSW DCTAMATERT FLQDFMGVSS 180 AGAAAAPTPW AITTREPETL PLFPVGGGGG GQECVGHGNL QRWGSAAAAA ATTTATAQQH 240 QLLQQHNFYS SSSSSNQLPS QDGSAAGTSL ELTLSSYCTP YPAGSSM |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription repressor required for the formation and development of tiller buds and panicles (PubMed:25697101). Required for tiller formation and female sterility (PubMed:27194802). Required for the early developmental stages of axillary meristem formation (PubMed:25841039). Plays a role in maintaining the axillary premeristem zone and in promoting the formation of the axillary meristem by promoting OSH1 expression (PubMed:25841039). Does not seem to be involved in maintenance of the shoot apical meristem (SAM) (PubMed:25841039). {ECO:0000269|PubMed:25697101, ECO:0000269|PubMed:25841039, ECO:0000269|PubMed:27194802}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00265 | DAP | Transfer from AT2G17950 | Download |
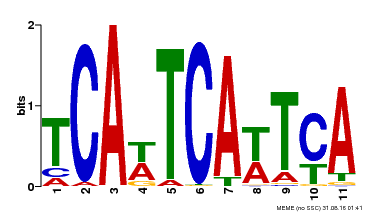
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | OB04G35540.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by the cytokinin 6-benzylaminopurine. {ECO:0000269|PubMed:17351053}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AB218894 | 1e-150 | AB218894.1 Oryza sativa Japonica Group mRNA for OsWUS protein, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006653851.1 | 0.0 | PREDICTED: WUSCHEL-related homeobox 1B-like | ||||
| Swissprot | Q7XM13 | 1e-117 | WOX1_ORYSJ; WUSCHEL-related homeobox 1 | ||||
| TrEMBL | J3M2C9 | 0.0 | J3M2C9_ORYBR; Uncharacterized protein | ||||
| STRING | OB04G35540.1 | 0.0 | (Oryza brachyantha) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP12465 | 31 | 36 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G17950.1 | 5e-35 | WOX family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 102720794 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



