 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OB06G10790.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 781aa MW: 88975.5 Da PI: 10.5473 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 100.7 | 9.9e-32 | 608 | 661 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
pr+rWt++LH+rFv+ave LGG+e+AtPk++lelm+vk+Ltl+hvkSHLQ+YR+
OB06G10790.1 608 PRMRWTTTLHARFVHAVELLGGHERATPKSVLELMDVKDLTLAHVKSHLQMYRT 661
8****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 4.84E-15 | 605 | 662 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 3.9E-28 | 606 | 661 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 4.6E-23 | 608 | 662 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 6.5E-7 | 609 | 660 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009944 | Biological Process | polarity specification of adaxial/abaxial axis | ||||
| GO:0048481 | Biological Process | plant ovule development | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 781 aa Download sequence Send to blast |
MELFPRQLDL SLNISTPSSS SSPSPAPAAT SWPWTSAKQL PADKEAAAMT ASARSAPLLP 60 RNSDVCSTRG AAAGAXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX 120 XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX 180 XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX 240 XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX 300 XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX 360 XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX 420 XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX XXXXXXXXXX 480 XXXXXXXXXX XXXXXXXTTP THHGGGLANL QKQQQQLKAA AMQQLKEAAM QQPQPIHGVP 540 VYHHQQQQQR RQLHQPHAVV GDRRSDGGGG RRLFSHVGVA TPRTLPSPSP SRSMLARLPP 600 GRRGVRAPRM RWTTTLHARF VHAVELLGGH ERATPKSVLE LMDVKDLTLA HVKSHLQMYR 660 TIKNTDRPVS YAGQANDGFD NAPGGDISDD SFTDGLLRQN RSMLASGEQN DTNIYSGLWS 720 NNTSGKVDGL DLGLPVSEPA NEFYRIYLKN AHRSVGLETS VLSLPGRPNL EFTLGIGKAS 780 Q |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4r_A | 1e-15 | 609 | 663 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 1e-15 | 609 | 663 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 1e-15 | 609 | 663 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 1e-15 | 609 | 663 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00174 | DAP | Transfer from AT1G32240 | Download |
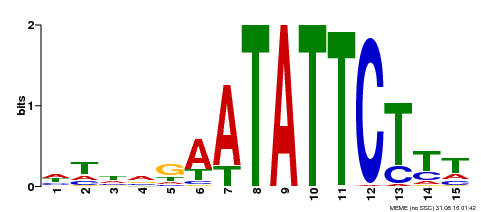
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | OB06G10790.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP17435 | 11 | 11 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G32240.1 | 1e-43 | G2-like family protein | ||||



