 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OB06G16250.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 712aa MW: 77963.6 Da PI: 6.3819 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 65.7 | 6.5e-21 | 16 | 71 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
r++ +++t+ q+++Le++F+++++p++++r++L+++lgL+ rq+k+WFqNrR+++k
OB06G16250.1 16 RKRYHRHTPRQIQQLEAMFKECPHPDENQRAQLSRELGLEPRQIKFWFQNRRTQMK 71
688899***********************************************998 PP
| |||||||
| 2 | START | 152.7 | 3.1e-48 | 212 | 441 | 3 | 206 |
HHHHHHHHHHHHHHC-TT-EEEE..........EXCCTTEEEEEEESSS.......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S....EE CS
START 3 aeeaaqelvkkalaeepgWvkss..........esengdevlqkfeeskv......dsgealrasgvvdmvlallveellddkeqWdetla....ka 79
a +a++el+++a+a++ W+ks+ e +n d++ ++f++ ++ ++e +r+sg+v m ++ l ++d++ +W e ++ ka
OB06G16250.1 212 ATRAMDELIRLAQAGDHIWSKSPgggvsggdsrETLNVDTYDSIFSKPGGsyrapsINVEGSRESGLVLMSAVALADVFMDTN-KWMEFFPsivsKA 307
6789***********************999999999999999999887779****99****************9999999999.************* PP
EEEEEECTT.....EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--.-TTSEE-EESSEEEEEEEECTCEEEEEEEE-E CS
START 80 etlevissg.....galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppesssvvRaellpSgiliepksnghskvtwvehv 170
t++v+ +g + l lm+ el ++p+vp R++ fvRy+rq ++g w+i+dvSvd +++ ++ R+++lpSg+li +++ng+skvtwveh+
OB06G16250.1 308 HTIDVLVNGmggrsESLILMYEELHIMTPAVPtREVSFVRYCRQIEQGLWAIADVSVDLQRDAHFGAPPPRSRRLPSGCLIADMANGYSKVTWVEHM 404
**************************************************************98899****************************** PP
E--SSXX.HHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 171 dlkgrlp.hwllrslvksglaegaktwvatlqrqcek 206
+++++ p l+r lv sg+a+ga +w+a+lqr ce+
OB06G16250.1 405 EVEEKNPiNVLYRDLVLSGAAFGAHRWLAALQRACER 441
*******999*************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.88E-20 | 9 | 76 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.9E-22 | 10 | 78 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 18.204 | 13 | 73 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 4.8E-20 | 14 | 77 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 5.62E-20 | 15 | 74 | No hit | No description |
| Pfam | PF00046 | 1.8E-18 | 16 | 71 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 48 | 71 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 45.358 | 201 | 444 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 6.73E-31 | 202 | 443 | No hit | No description |
| CDD | cd08875 | 1.23E-110 | 205 | 440 | No hit | No description |
| SMART | SM00234 | 1.8E-33 | 210 | 441 | IPR002913 | START domain |
| Pfam | PF01852 | 1.3E-40 | 212 | 441 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 4.4E-19 | 460 | 701 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009828 | Biological Process | plant-type cell wall loosening | ||||
| GO:0010091 | Biological Process | trichome branching | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 712 aa Download sequence Send to blast |
MDFGDEPEGS DSQRRRKRYH RHTPRQIQQL EAMFKECPHP DENQRAQLSR ELGLEPRQIK 60 FWFQNRRTQM KAQHERADNC FLRAENDKIR CENIAIREAL KNVICPTCGG PPVGEDYFDE 120 QKLRMENARL KEELDRVSNL TSKYLGRPFT QLPPATPMSV SSLDLSVGGM SGPGLGGPSL 180 DLDLLSGGSS GIPFQLPAPV SDMERPMMAD MATRAMDELI RLAQAGDHIW SKSPGGGVSG 240 GDSRETLNVD TYDSIFSKPG GSYRAPSINV EGSRESGLVL MSAVALADVF MDTNKWMEFF 300 PSIVSKAHTI DVLVNGMGGR SESLILMYEE LHIMTPAVPT REVSFVRYCR QIEQGLWAIA 360 DVSVDLQRDA HFGAPPPRSR RLPSGCLIAD MANGYSKVTW VEHMEVEEKN PINVLYRDLV 420 LSGAAFGAHR WLAALQRACE RYASLVALGV PHHIAGGAPT PEGKRSMMKL SQRMVNSFCS 480 SLGASQMHQW TTLSGSNEVS VRVTMHRSTD PGQPNGVVLS AATSIWLPVP CDHVFAFVRD 540 ENTRSQWDVL SHGNQVQEVS RIPNGSNPGN CISLLRVITL INLLHNGLNA SQNSMLILQE 600 SCTDASGSLV VYSPIDIPAA NVVMSGEDPS SIPLLPSGFT ILPDXXXXXX XXXXXXXXXX 660 VGSVVTVAFQ ILVSSLPSSK LNAESVATVN GLITTTVEQI KAALNCSAHG HP |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
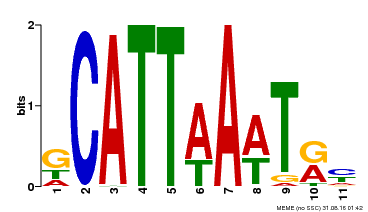
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00232 | DAP | Transfer from AT1G73360 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | OB06G16250.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015642098.1 | 0.0 | homeobox-leucine zipper protein ROC8 | ||||
| Swissprot | Q69T58 | 0.0 | ROC8_ORYSJ; Homeobox-leucine zipper protein ROC8 | ||||
| TrEMBL | J3MC81 | 0.0 | J3MC81_ORYBR; Uncharacterized protein | ||||
| STRING | OB06G16250.1 | 0.0 | (Oryza brachyantha) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2912 | 37 | 70 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73360.1 | 0.0 | homeodomain GLABROUS 11 | ||||



