 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OB06G25740.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 347aa MW: 36407 Da PI: 7.8087 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 154 | 1.1e-47 | 11 | 156 | 2 | 149 |
DUF822 2 gsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpl..eeaeaagssasaspesslqs 96
g++r ptw+ErEnn+rRERr a G a+Gny+lpk++DnneVlkALc+eAGw+ve+DGttyrkg+kp+ e+++ +g+sas sp+ss+q
OB06G25740.1 11 GGTRVPTWRERENNRRRERRGGG-GA---RGASAYGNYNLPKHCDNNEVLKALCNEAGWTVEPDGTTYRKGCKPPpsERPDPIGRSASPSPCSSYQ- 102
789***************98765.44...4999*****************************************9999******************. PP
DUF822 97 slkssalaspvesysaspksssfpspssldsislasa........asllpvlsvlslvsss 149
+sp+ sy++sp+sssfps+ s+++i+la + +sl+p+l++l+l+ss
OB06G25740.1 103 -------PSPRGSYNPSPASSSFPSSGSSSHITLAGNnliggvegSSLIPWLKTLPLSSSY 156
.......9*************************999899****999*******99986664 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 1.2E-57 | 11 | 150 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 347 aa Download sequence Send to blast |
XXXXGAGGGL GGTRVPTWRE RENNRRRERR GGGGARGASA YGNYNLPKHC DNNEVLKALC 60 NEAGWTVEPD GTTYRKGCKP PPSERPDPIG RSASPSPCSS YQPSPRGSYN PSPASSSFPS 120 SGSSSHITLA GNNLIGGVEG SSLIPWLKTL PLSSSYASSS KFPQLHHLYF NGGSISAPVT 180 PPSSSPTRTP RMRTDWENAS VQPPWASANY TSLPNSTPPS PGHQIAPDPA WLAGFQISSA 240 GPSSPTYNLV SPNPFGVFKE AIASTSRVCT PGQSGTCSPV MGGVPAHHDV QMVDGAPDDF 300 AFGSSSNGKN ESPGLVKAWE GERIHEECAS DELELTLGSS KTRADPS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5zd4_A | 6e-19 | 37 | 84 | 399 | 446 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_B | 6e-19 | 37 | 84 | 399 | 446 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_C | 6e-19 | 37 | 84 | 399 | 446 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_D | 6e-19 | 37 | 84 | 399 | 446 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | May function in brassinosteroid signaling. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00248 | DAP | Transfer from AT1G78700 | Download |
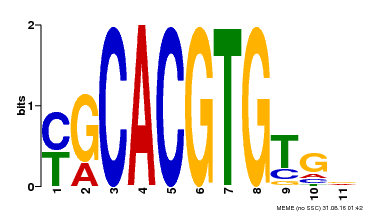
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | OB06G25740.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK101229 | 0.0 | AK101229.1 Oryza sativa Japonica Group cDNA clone:J033031H19, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015693609.1 | 0.0 | PREDICTED: LOW QUALITY PROTEIN: protein BZR1 homolog 3-like | ||||
| Swissprot | Q5Z9E5 | 0.0 | BZR3_ORYSJ; Protein BZR1 homolog 3 | ||||
| TrEMBL | J3MEY0 | 0.0 | J3MEY0_ORYBR; Uncharacterized protein | ||||
| STRING | OB06G25740.1 | 0.0 | (Oryza brachyantha) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1366 | 38 | 112 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G78700.1 | 9e-66 | BES1/BZR1 homolog 4 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



