 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OB06G31100.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 425aa MW: 45043.5 Da PI: 9.1313 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 131.2 | 3.7e-41 | 167 | 243 | 1 | 77 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S-- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkq 77
+Cq+egC+adls ak+yhrrhkvCe h+ka+vv++ g++qrfCqqCsrfh l+efDe+krsCr+rLa+hn+rrrk++
OB06G31100.1 167 RCQAEGCKADLSGAKHYHRRHKVCEYHAKASVVAAGGKQQRFCQQCSRFHVLTEFDEAKRSCRKRLAEHNRRRRKPA 243
6**************************************************************************87 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 3.8E-35 | 160 | 229 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.496 | 165 | 242 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 6.67E-39 | 166 | 245 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 3.9E-31 | 168 | 241 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009554 | Biological Process | megasporogenesis | ||||
| GO:0009556 | Biological Process | microsporogenesis | ||||
| GO:0048653 | Biological Process | anther development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 425 aa Download sequence Send to blast |
MMSGRMNASG EEAFPFGAMQ PGGPYVGFEH GAAAVAAAAQ RAGMQQHHHH HMYDSLDFAA 60 AMQFQEPQLL ALPPSMPMPP PPMPMPLQMP VPMPGDVYPA LGIVKREGGG VGAQDGSAAA 120 GRIGLNLGRR TYFSPGDMLA VDRLLMRSRL GGVFGLGFGG AHHQPPRCQA EGCKADLSGA 180 KHYHRRHKVC EYHAKASVVA AGGKQQRFCQ QCSRFHVLTE FDEAKRSCRK RLAEHNRRRR 240 KPATTAAAAS GKDAAASPVA AGKKPSGATT SSYTGDSKNV VAMSAAKSPI SSNTSVISCL 300 AEQQAGKQAA RPTALTLGAA PPPHDREVQQ LGAVLHAQHH HHHHHHHHQQ QEHIQVSSLV 360 HIAGNNNNNH QNGGGGGSNI LSCSSVCSSA LPSTATNGEV SDQNNDNSHN NGGNNNMHLF 420 EVDFM |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 3e-29 | 168 | 241 | 11 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 224 | 241 | KRSCRKRLAEHNRRRRKP |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00090 | SELEX | Transfer from AT1G02065 | Download |
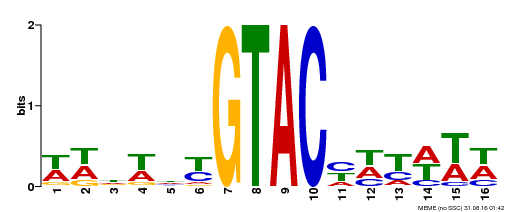
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | OB06G31100.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AP005760 | 1e-172 | AP005760.3 Oryza sativa Japonica Group genomic DNA, chromosome 6, BAC clone:B1047G05. | |||
| GenBank | AP014962 | 1e-172 | AP014962.1 Oryza sativa Japonica Group DNA, chromosome 6, cultivar: Nipponbare, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006656320.2 | 0.0 | PREDICTED: squamosa promoter-binding-like protein 10 | ||||
| Swissprot | A2YFT9 | 1e-151 | SPL10_ORYSI; Squamosa promoter-binding-like protein 10 | ||||
| Swissprot | Q0DAE8 | 1e-151 | SPL10_ORYSJ; Squamosa promoter-binding-like protein 10 | ||||
| TrEMBL | J3MGG6 | 0.0 | J3MGG6_ORYBR; Uncharacterized protein | ||||
| STRING | OB06G31100.1 | 0.0 | (Oryza brachyantha) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2375 | 35 | 93 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G02065.1 | 3e-45 | squamosa promoter binding protein-like 8 | ||||



