 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OB07G25720.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 192aa MW: 20504.8 Da PI: 4.8233 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 30.2 | 1.5e-09 | 2 | 56 | 89 | 145 |
DUF822 89 spesslq.sslkssalaspvesysaspksssfpspssldsislasaasllpvlsvlsl 145
sp+ss+q +s+ ss+++spv+sy+asp+sssfpsps+ld++s ++llp+l+ l++
OB07G25720.1 2 SPCSSTQlLSAPSSSFPSPVPSYHASPASSSFPSPSRLDNASP---SCLLPFLRGLPN 56
89*****9*******************************9854...688888877665 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 1.4E-6 | 1 | 42 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009742 | Biological Process | brassinosteroid mediated signaling pathway | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005829 | Cellular Component | cytosol | ||||
| GO:0001046 | Molecular Function | core promoter sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 192 aa Download sequence Send to blast |
MSPCSSTQLL SAPSSSFPSP VPSYHASPAS SSFPSPSRLD NASPSCLLPF LRGLPNLPPL 60 RVSSSAPIRK PDWDVDPFRH PFFAVSAPAS PTRGRRLEHP DTIPECDESD VSTVDSGRWI 120 SFQMATTAPT SPTYNLVNPG ASNSNSMEIE GAAGRGGPEF EFDKGRVTPW EGERIHEVAA 180 EELELTLGVG AK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Positive brassinosteroid-signaling protein. Mediates downstream brassinosteroid-regulated growth response and feedback inhibition of brassinosteroid biosynthetic genes. May act as transcriptional repressor by binding the brassinosteroid-response element (5'-CGTGCG-3') in the promoter of GRAS32 (AC Q9LWU9), another positive regulator of brassinosteroid signaling (By similarity). {ECO:0000250}. | |||||
| UniProt | Positive brassinosteroid-signaling protein. Mediates downstream brassinosteroid-regulated growth response and feedback inhibition of brassinosteroid (BR) biosynthetic genes (PubMed:17699623, PubMed:19220793). May act as transcriptional repressor by binding the brassinosteroid-response element (BREE) (5'-CGTG(T/C)G-3') in the promoter of DLT (AC Q9LWU9), another positive regulator of BR signaling (PubMed:19220793). Acts as transcriptional repressor of LIC, a negative regulator of BR signaling, by binding to the BRRE element of its promoter. BZR1 and LIC play opposite roles in BR signaling and regulation of leaf bending (PubMed:22570626). {ECO:0000269|PubMed:17699623, ECO:0000269|PubMed:19220793, ECO:0000269|PubMed:22570626}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00073 | ChIP-chip | Transfer from AT1G19350 | Download |
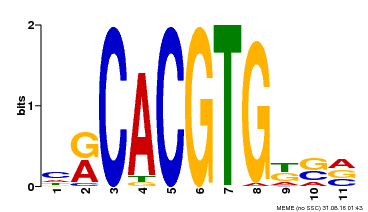
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | OB07G25720.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated by 24-epibrassinolide. {ECO:0000269|PubMed:22570626}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK106748 | 1e-166 | AK106748.1 Oryza sativa Japonica Group cDNA clone:002-115-C06, full insert sequence. | |||
| GenBank | AP004276 | 1e-166 | AP004276.3 Oryza sativa Japonica Group genomic DNA, chromosome 7, PAC clone:P0453G03. | |||
| GenBank | AP014963 | 1e-166 | AP014963.1 Oryza sativa Japonica Group DNA, chromosome 7, cultivar: Nipponbare, complete sequence. | |||
| GenBank | CP012615 | 1e-166 | CP012615.1 Oryza sativa Indica Group cultivar RP Bio-226 chromosome 7 sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006658737.1 | 1e-110 | PREDICTED: protein BZR1 homolog 1 | ||||
| Swissprot | B8B7S5 | 1e-108 | BZR1_ORYSI; Protein BZR1 homolog 1 | ||||
| Swissprot | Q7XI96 | 1e-108 | BZR1_ORYSJ; Protein BZR1 homolog 1 | ||||
| TrEMBL | J3MME0 | 1e-135 | J3MME0_ORYBR; Uncharacterized protein | ||||
| STRING | OB07G25720.1 | 1e-135 | (Oryza brachyantha) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP5474 | 34 | 53 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G19350.6 | 3e-41 | BES1 family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



