 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OB09G16960.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 487aa MW: 52954.1 Da PI: 9.7451 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 92.6 | 2.9e-29 | 137 | 196 | 1 | 59 |
---SS-EEEEEEE--TT-SS-EEEEEE-ST.T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 1 ldDgynWrKYGqKevkgsefprsYYrCtsa.gCpvkkkversaedpkvveitYegeHnhe 59
++Dgy+WrKYGqK+ ++++ pr+Y+rC++a +Cpvkkkv+rsaed ++++ tYegeHnh+
OB09G16960.1 137 VKDGYQWRKYGQKVTRDNPSPRAYFRCAFApSCPVKKKVQRSAEDSSLLVATYEGEHNHP 196
58***************************99****************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50811 | 27.473 | 132 | 198 | IPR003657 | WRKY domain |
| Gene3D | G3DSA:2.20.25.80 | 3.5E-28 | 135 | 198 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 4.32E-25 | 135 | 198 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.4E-34 | 137 | 197 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 5.3E-24 | 138 | 196 | IPR003657 | WRKY domain |
| Pfam | PF00954 | 1.9E-8 | 383 | 467 | IPR000858 | S-locus glycoprotein domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0002237 | Biological Process | response to molecule of bacterial origin | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0048544 | Biological Process | recognition of pollen | ||||
| GO:0050691 | Biological Process | regulation of defense response to virus by host | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 487 aa Download sequence Send to blast |
MDTAWRGGGR SPVCLDLCVG LSPAKILEAK VTQMSEENRR LTEVITRLYG SHVARLGLDD 60 AASPQRRPVS PLSSRKRGRE SMETANSCDG NGNGQKSGAA AEADHAESFA MDDGTCRRIK 120 VSRVCRRIDP SDATLVVKDG YQWRKYGQKV TRDNPSPRAY FRCAFAPSCP VKKKVQRSAE 180 DSSLLVATYE GEHNHPLPSR PGELAAGGGG GVGSAGSLPC SISINPSGPT IALDLTKNGG 240 AVQVANRDTP LNGTAGVLVI NGLGVLFLLD SSGQGRMVVV EHDSSWHGVF IISGGGGTAA 300 RVREPCRARP EQQQLAVVVI RSLIEHPIAD MRLDLDTNLQ TGAKWFLTSW RAPDDPATRE 360 YRRVLEKKGL PDCVSWRGAT KKYRMGAWNG LWFSGVPKMA FPSSSVFGNQ EVDRPNKMAF 420 IFNPFAGGPL SRLVLNEAGI AQRLVWDPSS KAWNAYAQTP HKITVAYEHE MLMSWLIHGR 480 SDIRTKH |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2ayd_A | 2e-21 | 136 | 202 | 13 | 76 | WRKY transcription factor 1 |
| 5gyy_A | 3e-19 | 242 | 460 | 56 | 263 | S-receptor kinase SRK9 |
| 5gyy_B | 3e-19 | 242 | 460 | 56 | 263 | S-receptor kinase SRK9 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription repressor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element. Regulates, probably indirectly, the activation of defense-related genes during defense response (By similarity). Modulates plant innate immunity against X.oryzae pv. oryzae (Xoo) (By similarity). {ECO:0000250|UniProtKB:Q6EPZ2, ECO:0000250|UniProtKB:Q6QHD1}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00251 | DAP | Transfer from AT1G80840 | Download |
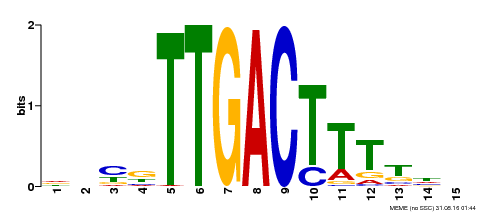
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | OB09G16960.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by biotic elicitors (e.g. fungal chitin oligosaccharide) (By similarity). Induced by pathogen infection (e.g. M.grisea and X.oryzae pv. oryzae (Xoo)) (PubMed:16528562). Accumulates after treatment with benzothiadiazole (BTH) and salicylic acid (SA) (By similarity). {ECO:0000250|UniProtKB:Q6EPZ2, ECO:0000269|PubMed:16528562}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015696342.1 | 0.0 | PREDICTED: uncharacterized protein LOC102714490 | ||||
| Swissprot | Q6IEK5 | 1e-130 | WRK76_ORYSI; WRKY transcription factor WRKY76 | ||||
| TrEMBL | J3MXG7 | 0.0 | J3MXG7_ORYBR; Uncharacterized protein | ||||
| STRING | OB09G16960.1 | 0.0 | (Oryza brachyantha) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6660 | 35 | 52 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G80840.1 | 2e-46 | WRKY DNA-binding protein 40 | ||||



