 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OB09G21280.1 | ||||||||
| Common Name | LOC102711094 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | EIL | ||||||||
| Protein Properties | Length: 611aa MW: 68158.6 Da PI: 5.8056 | ||||||||
| Description | EIL family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | EIN3 | 432.2 | 4.9e-132 | 39 | 418 | 1 | 353 |
XXXXXXXXXXXXXXXXXXXXXXX..XXXXX.XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX CS
EIN3 1 eelkkrmwkdqmllkrlkerkkqlledkeaatgakksnksneqarrkkmsraQDgiLkYMlkemevcnaqGfvYgiipekgkpvegasdsLraWWke 97
e+l +rmwkd+++l+r+ker+ l + + a+++++k + +++qa rkkmsra DgiLkYMlk mevcna+GfvYgiip+kgkpv+gasd++raWWke
OB09G21280.1 39 EDLARRMWKDRVKLRRIKERQHTL-ALQLAELDKSKPKAISDQAMRKKMSRAHDGILKYMLKLMEVCNARGFVYGIIPDKGKPVSGASDNIRAWWKE 134
89*******************985.578888****************************************************************** PP
XXXXXXXXXXXXXXXXXXXXXXXXXXXX....XX----STTS-HHHHHHHHHHHSSSSSS-TTS--TTT--HHHH---S--HHHHHHT--TT--.-- CS
EIN3 98 kvefdrngpaaiskyqaknlilsgesslqtersseshslselqDTtlgSLLsalmqhcdppqrrfplekgvepPWWPtGkelwwgelglskdqgtpp 194
kv+fd+ngpaai+ky+++n++++++ s+ + +++hsl++lqD+tlgSLLs+lmqhc+ppqr++plekg++pPWWP+G+e+ww +lgl++ q pp
OB09G21280.1 135 KVKFDKNGPAAIAKYESENMASADAPSS---GIKNQHSLMDLQDATLGSLLSSLMQHCNPPQRKYPLEKGTPPPWWPSGDEDWWIALGLPRGQI-PP 227
*********************9988888...88************************************************************9.9* PP
---GGG--HHHHHHHHHHHHHHTGGGHHHHHHTTTTSSSSTTT--SHHHHHHHHHHTTTTT-S--XXXXXX....XXXXXXXXXXXXXXXXXXXXXX CS
EIN3 195 ykkphdlkkawkvsvLtavikhmsptieeirelerqskylqdkmsakesfallsvlnqeekecatvsahss....slrkqspkvtlsceqkedvegk 287
ykkphdlkk+wkv+vLtavikhmsp++++ir+++r+sk+lqdkm+akes ++lsvl++ee+++ ++ +s ++++ t+s+++++dv+g
OB09G21280.1 228 YKKPHDLKKVWKVGVLTAVIKHMSPDFDKIRNHVRKSKCLQDKMTAKESLIWLSVLQREERLVYSIDNGMSevthLALEDRNGDTHSSNNEYDVDGI 324
***************************************************************999999554654333488899999********99 PP
XXXX.XXXXXXXXXX.................................XXXXXXXXXXXXXXXXXXXXX......XXXXXXX.XXXXXXXXXXXXXX CS
EIN3 288 keskikhvqavktta.................................gfpvvrkrkkkpsesakvsskevsrtcqssqfrgsetelifadknsisq 351
+e ++++ +++++ g++ krk+++++ ++v ++ev + + +++++i++d+n+++q
OB09G21280.1 325 EEAPLSTSSRDDEHDlplavvqlseehvptrreranvkrpnqvvpkkaGTKEPPKRKRPRHSATAV-EHEV---QRAVDAP-ENSRNIIPDMNRLDQ 416
999999999999988*********************************9*******9665555555.5545...4555555.57899******9998 PP
XX CS
EIN3 352 ne 353
e
OB09G21280.1 417 VE 418
65 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF04873 | 2.8E-125 | 39 | 287 | No hit | No description |
| Gene3D | G3DSA:1.10.3180.10 | 1.3E-70 | 161 | 294 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| SuperFamily | SSF116768 | 3.27E-61 | 167 | 291 | IPR023278 | Ethylene insensitive 3-like protein, DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0042762 | Biological Process | regulation of sulfur metabolic process | ||||
| GO:0071281 | Biological Process | cellular response to iron ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 611 aa Download sequence Send to blast |
MMDHLAIIAT ELGDSSDFEV EGIQNLTEND VSDEEIEAED LARRMWKDRV KLRRIKERQH 60 TLALQLAELD KSKPKAISDQ AMRKKMSRAH DGILKYMLKL MEVCNARGFV YGIIPDKGKP 120 VSGASDNIRA WWKEKVKFDK NGPAAIAKYE SENMASADAP SSGIKNQHSL MDLQDATLGS 180 LLSSLMQHCN PPQRKYPLEK GTPPPWWPSG DEDWWIALGL PRGQIPPYKK PHDLKKVWKV 240 GVLTAVIKHM SPDFDKIRNH VRKSKCLQDK MTAKESLIWL SVLQREERLV YSIDNGMSEV 300 THLALEDRNG DTHSSNNEYD VDGIEEAPLS TSSRDDEHDL PLAVVQLSEE HVPTRRERAN 360 VKRPNQVVPK KAGTKEPPKR KRPRHSATAV EHEVQRAVDA PENSRNIIPD MNRLDQVEIP 420 GMANQLTSFN EEGNTSEAFQ HRGNTQGQTH LPGADFNHYG NAQAANATPV SICMGGQPVP 480 YESSDNSRSK TGNIFPLDSD SGFNNLPSSY QTVPPKQSLP LSIMDHHVVP MGIRTPADNS 540 PYGDQIIGSG NSTSVPGDMQ LIDYPFYGEQ DKFAGSSFAG LPLDYISISS PIPDIDDLLL 600 HDDDLMEYLG T |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wij_A | 2e-66 | 167 | 290 | 9 | 132 | ETHYLENE-INSENSITIVE3-like 3 protein |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in the ethylene response pathway. {ECO:0000269|PubMed:9215635, ECO:0000269|PubMed:9851977}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00649 | PBM | Transfer from LOC_Os09g31400 | Download |
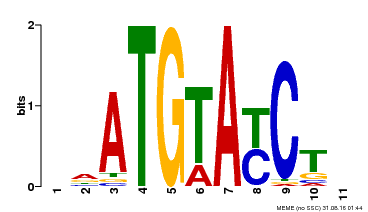
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | OB09G21280.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC108758 | 0.0 | AC108758.2 Oryza sativa (japonica cultivar-group) chromosome 9 BAC clone OSJNBa0066E19, complete sequence. | |||
| GenBank | AK065979 | 0.0 | AK065979.1 Oryza sativa Japonica Group cDNA clone:J013047J08, full insert sequence. | |||
| GenBank | AP014965 | 0.0 | AP014965.1 Oryza sativa Japonica Group DNA, chromosome 9, cultivar: Nipponbare, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006660770.1 | 0.0 | PREDICTED: ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Refseq | XP_015696822.1 | 0.0 | PREDICTED: ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| Swissprot | O23116 | 1e-147 | EIL3_ARATH; ETHYLENE INSENSITIVE 3-like 3 protein | ||||
| TrEMBL | J3MYP9 | 0.0 | J3MYP9_ORYBR; Uncharacterized protein | ||||
| STRING | OB09G21280.1 | 0.0 | (Oryza brachyantha) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3720 | 37 | 79 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73730.1 | 1e-149 | ETHYLENE-INSENSITIVE3-like 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 102711094 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



