 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OB10G25650.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 260aa MW: 28895.7 Da PI: 5.348 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 43.3 | 8.2e-14 | 36 | 80 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT+eE+ +++ + ++lG+g+W+ I+r + Rt+ q+ s+ qky
OB10G25650.1 36 PWTEEEHRRFLLGLQKLGKGDWRGISRNFVVSRTPTQVASHAQKY 80
8*******************************************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 18.253 | 29 | 85 | IPR017930 | Myb domain |
| SMART | SM00717 | 4.7E-10 | 33 | 83 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 2.92E-16 | 34 | 86 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 7.3E-19 | 35 | 84 | IPR006447 | Myb domain, plants |
| Gene3D | G3DSA:1.10.10.60 | 1.8E-11 | 35 | 79 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 3.5E-11 | 36 | 80 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.24E-9 | 36 | 81 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000122 | Biological Process | negative regulation of transcription from RNA polymerase II promoter | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0030307 | Biological Process | positive regulation of cell growth | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:2000469 | Biological Process | negative regulation of peroxidase activity | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 260 aa Download sequence Send to blast |
MVQQLLLLQP LLAAYMPSGH IMCAMFLTKV NFTGVPWTEE EHRRFLLGLQ KLGKGDWRGI 60 SRNFVVSRTP TQVASHAQKY FIRQANMTRR KRRSSLFDMV PDESMDLPPL PGSQEPETQV 120 LNQPALPPPR EEEEEVDSME SDTSAIAESS SASAVVPENL QPTYPVIVPA YFSPFLQFSV 180 PFWQNQKDED GGAQETHEIV KPVPVHSKSP INVDELVGMS KLSIGESNQE TVSTSLSLNL 240 VGGQNRQSAF HANPPTRAQA |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription repressor that binds to 5'-TATCCA-3' elements in gene promoters. Contributes to the sugar-repressed transcription of promoters containing SRS or 5'-TATCCA-3' elements. Transcription repressor involved in a cold stress response pathway that confers cold tolerance. Suppresses the DREB1-dependent signaling pathway under prolonged cold stress. DREB1 responds quickly and transiently while MYBS3 responds slowly to cold stress. They may act sequentially and complementarily for adaptation to short- and long-term cold stress (PubMed:20130099). {ECO:0000269|PubMed:12172034, ECO:0000269|PubMed:20130099}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00551 | DAP | Transfer from AT5G47390 | Download |
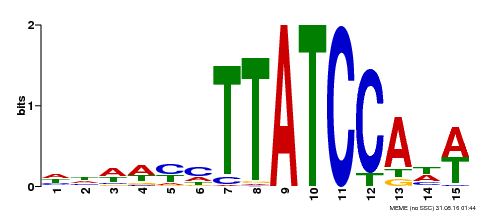
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | OB10G25650.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by sucrose and gibberellic acid (GA) (PubMed:12172034). Induced by cold stress in roots and shoots. Induced by salt stress in shoots. Down-regulated by abscisic aci (ABA) in shoots (PubMed:20130099). {ECO:0000269|PubMed:12172034, ECO:0000269|PubMed:20130099}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK059716 | 0.0 | AK059716.1 Oryza sativa Japonica Group cDNA clone:001-032-G03, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015614288.1 | 1e-150 | transcription factor MYBS3 | ||||
| Swissprot | Q7XC57 | 1e-151 | MYBS3_ORYSJ; Transcription factor MYBS3 | ||||
| TrEMBL | J3N4W7 | 0.0 | J3N4W7_ORYBR; Uncharacterized protein | ||||
| STRING | OB10G25650.1 | 0.0 | (Oryza brachyantha) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1939 | 37 | 91 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G16350.1 | 3e-44 | MYB_related family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



