 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OBART01G08110.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 959aa MW: 104762 Da PI: 4.9715 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 46.8 | 7.1e-15 | 66 | 105 | 8 | 48 |
HHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 8 EdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
Ede++v+ v++lG++ W+tIa+ ++ gR +kqc++rw+++l
OBART01G08110.2 66 EDEIIVQMVNKLGPKKWSTIAQALP-GRIGKQCRERWYNHL 105
9************************.*************97 PP
| |||||||
| 2 | Myb_DNA-binding | 48.6 | 1.9e-15 | 111 | 153 | 1 | 45 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwq 45
+ +WT+eE+ l++a++++G++ W+ + ++ gRt++++k++w+
OBART01G08110.2 111 KEAWTQEEEITLIHAHRMYGNK-WAELTKFLP-GRTDNSIKNHWN 153
579*******************.*********.***********8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 13.227 | 56 | 105 | IPR017930 | Myb domain |
| SMART | SM00717 | 3.0E-10 | 58 | 107 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 8.0E-22 | 66 | 112 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 1.5E-12 | 66 | 105 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.81E-12 | 66 | 105 | No hit | No description |
| SuperFamily | SSF46689 | 1.46E-27 | 85 | 162 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 24.107 | 106 | 160 | IPR017930 | Myb domain |
| SMART | SM00717 | 2.2E-14 | 110 | 158 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 6.2E-14 | 111 | 154 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.96E-11 | 113 | 153 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 1.1E-21 | 113 | 160 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0032875 | Biological Process | regulation of DNA endoreduplication | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003713 | Molecular Function | transcription coactivator activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 959 aa Download sequence Send to blast |
MESLLDLRRL LKKEKSAMNQ KGDDLSMGGP PVRHGVPPKE IGPLKRMPYC PQLFRHTTGR 60 TGKKLEDEII VQMVNKLGPK KWSTIAQALP GRIGKQCRER WYNHLNPGIN KEAWTQEEEI 120 TLIHAHRMYG NKWAELTKFL PGRTDNSIKN HWNSSVKKKV NSYMSSGLLT QVSCLPLNEY 180 SANCNSSPAM TQQNSEDSGC FAVREVENSS GCSQSSLAKV SCSQVHDTTV PLGCDLQVNA 240 NFDKNEAHDS QSSMGPQACY TSAEAVASAL PAVHCHVSSS NLDPDQHLQE DFAQGLNLDM 300 TIDEMPTVPS VADNQTVCSI ENHERSLEPY DVAMEVPLSM LSSDSGAEQK LHFMSEADFN 360 SPNCLKSELW QDISLQGLLS GPDAVEADSF SRSNHQLDVY SSEADTHFLA PPYMPQTSNS 420 SSVMGLADDQ SPQMSVPPSL ICSNAMTDDA PFDNRPGRKE MPLSQAEVVT QSSSSSGDAE 480 MFANPGCSND RHVPSSTMES IPECGDQQVT NAEEPEASLE KEPSLTQSVT APDEQDKGAL 540 YYEPPRFPSL DVPFVSCDLV TSGDLQEFSP LGIRQLMHST MNVCTPMRLW GSPTHDESTG 600 VLLKSAAKSF ICTPSILKKR HRDLLSPIPD KRIEKKYGTE KDRGVSDTSS TGIQTSCINA 660 TKDDALITTV LRIERSASSK SLEKKLVFSD ENKENLGYTT EQTKDGKSAG NDEHMDEQTT 720 GERSSATNVA TNADLSGNLQ PAGILIEHSG DDPISSDYGK NTMNQKLNTN VKSLSVCKEG 780 VCAKSKPTEL IVEKSSPCIN VDYEYVNILA DTPGIKRGLE SPSAWKSPWF VDMHFQGSYF 840 TSPADSYDAL GLMKQINVQT AAALVEAREV LASGGQCDNI SSDKENTGNP DAKKEPGTTK 900 LQTKIMAEGR VLDFECTTPV RSSDKNAGSN LGRYLSSPIP SSHLLKSEDF SRLFTRKVG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1mse_C | 3e-40 | 64 | 160 | 9 | 105 | C-Myb DNA-Binding Domain |
| 1msf_C | 3e-40 | 64 | 160 | 9 | 105 | C-Myb DNA-Binding Domain |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00505 | DAP | Transfer from AT5G11510 | Download |
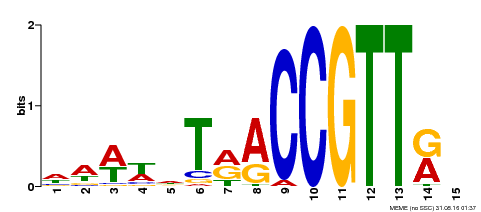
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK318601 | 0.0 | AK318601.1 Oryza sativa Japonica Group cDNA, clone: J100066E24, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015625869.1 | 0.0 | transcription factor MYB3R-1 | ||||
| TrEMBL | A0A0D3ELC5 | 0.0 | A0A0D3ELC5_9ORYZ; Uncharacterized protein | ||||
| STRING | ORGLA01G0073200.1 | 0.0 | (Oryza glaberrima) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G11510.1 | 3e-89 | myb domain protein 3r-4 | ||||



