 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OBART01G11630.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | TALE | ||||||||
| Protein Properties | Length: 259aa MW: 28791.5 Da PI: 7.0207 | ||||||||
| Description | TALE family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 28.9 | 2e-09 | 193 | 226 | 22 | 55 |
SSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHH CS
Homeobox 22 nrypsaeereeLAkklgLterqVkvWFqNrRake 55
+yp++e++ +LA+++gL+ +q+ +WF N+R ++
OBART01G11630.1 193 WPYPTEEDKLRLAARTGLDPKQINNWFINQRKRH 226
69*****************************885 PP
| |||||||
| 2 | ELK | 33.4 | 9.7e-12 | 146 | 167 | 1 | 22 |
ELK 1 ELKhqLlrKYsgyLgsLkqEFs 22
ELK++Ll+KYsg+L+ L++EF+
OBART01G11630.1 146 ELKEMLLKKYSGCLSRLRSEFL 167
9********************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01255 | 2.4E-19 | 1 | 43 | IPR005540 | KNOX1 |
| Pfam | PF03790 | 1.1E-20 | 1 | 41 | IPR005540 | KNOX1 |
| SMART | SM01256 | 2.5E-28 | 49 | 100 | IPR005541 | KNOX2 |
| Pfam | PF03791 | 1.2E-24 | 52 | 99 | IPR005541 | KNOX2 |
| Pfam | PF03789 | 1.1E-8 | 146 | 167 | IPR005539 | ELK domain |
| PROSITE profile | PS51213 | 10.423 | 146 | 166 | IPR005539 | ELK domain |
| SMART | SM01188 | 3.6E-6 | 146 | 167 | IPR005539 | ELK domain |
| PROSITE profile | PS50071 | 12.843 | 166 | 229 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 3.25E-19 | 168 | 231 | IPR009057 | Homeodomain-like |
| SMART | SM00389 | 1.1E-13 | 168 | 233 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 5.0E-27 | 171 | 229 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 1.94E-13 | 178 | 230 | No hit | No description |
| Pfam | PF05920 | 5.1E-17 | 186 | 225 | IPR008422 | Homeobox KN domain |
| PROSITE pattern | PS00027 | 0 | 204 | 227 | IPR017970 | Homeobox, conserved site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010073 | Biological Process | meristem maintenance | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 259 aa Download sequence Send to blast |
MKAQIAGHPR YPTLLSAYIE CRKVGAPPEV ASLLEEIGRE RRAGGGGGGA GQIGVDPELD 60 EFMEAYCRVL VRYKEELSRP FDEAASFLSS IQTQLSNLCS GATSPPATTA THSDEMVGSS 120 DEDQCSGETD MLDIGQEQSS RLADHELKEM LLKKYSGCLS RLRSEFLKKR KKGKLPKDAR 180 SALLEWWNTH YRWPYPTEED KLRLAARTGL DPKQINNWFI NQRKRHWKPS DGMRFALMEG 240 VAGGSSGTTL YFDTGTIGP |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 161 | 170 | LRSEFLKKRK |
| 2 | 167 | 171 | KKRKK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in shoot formation during early embryogenesis. {ECO:0000269|PubMed:10488233}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00670 | PBM | Transfer from GRMZM2G087741 | Download |
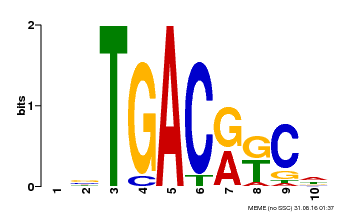
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AB028883 | 0.0 | AB028883.1 Oryza sativa mRNA for knotted1-type homeobox protein OSH6, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015634297.1 | 0.0 | homeobox protein knotted-1-like 1 | ||||
| Swissprot | Q9FP29 | 0.0 | KNOS1_ORYSJ; Homeobox protein knotted-1-like 1 | ||||
| TrEMBL | A0A0D9Y7A2 | 0.0 | A0A0D9Y7A2_9ORYZ; Uncharacterized protein | ||||
| TrEMBL | I1NM88 | 0.0 | I1NM88_ORYGL; Uncharacterized protein | ||||
| STRING | OGLUM01G14210.1 | 0.0 | (Oryza glumipatula) | ||||
| STRING | ORGLA01G0097800.1 | 0.0 | (Oryza glaberrima) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1698 | 38 | 92 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G23380.1 | 1e-78 | KNOTTED1-like homeobox gene 6 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



