 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OBART03G05300.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 363aa MW: 39422.5 Da PI: 6.9045 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 56.5 | 4.6e-18 | 130 | 183 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k+++++ eq+++Le+ Fe +++ e++ +LA+ lgL+ rqV +WFqNrRa++k
OBART03G05300.1 130 KKRRLNVEQVRTLEKNFELGNKLEPERKMQLARALGLQPRQVAIWFQNRRARWK 183
4568999**********************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 128.9 | 2e-41 | 129 | 220 | 1 | 92 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreelk 92
ekkrrl+ eqv++LE++Fe +kLeperK++lar+Lglqprqva+WFqnrRAR+ktkqlEkdy+aLkr++da+k+en++L +++++L++e+
OBART03G05300.1 129 EKKRRLNVEQVRTLEKNFELGNKLEPERKMQLARALGLQPRQVAIWFQNRRARWKTKQLEKDYDALKRQLDAVKAENDALLNHNKKLQAEIV 220
69**************************************************************************************9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 1.5E-19 | 122 | 187 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.41 | 125 | 185 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.2E-17 | 128 | 189 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 9.35E-17 | 130 | 186 | No hit | No description |
| Pfam | PF00046 | 2.1E-15 | 130 | 183 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 8.1E-19 | 132 | 193 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 1.3E-5 | 156 | 165 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 160 | 183 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 1.3E-5 | 165 | 181 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 4.8E-14 | 185 | 225 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009744 | Biological Process | response to sucrose | ||||
| GO:0048826 | Biological Process | cotyledon morphogenesis | ||||
| GO:0080022 | Biological Process | primary root development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 363 aa Download sequence Send to blast |
MASNGMASSP SSFFPPNFLL HMAQQQAAPP HDPQEHHHHH HHGHHGHHHE QQQQQQHHHH 60 LGPPPPPPPH PHNPFLPSSA QCPSLQEFRG MAPMLGKRPM SYGDGGGGGD EVNGGGEDEL 120 SDDGSQAGEK KRRLNVEQVR TLEKNFELGN KLEPERKMQL ARALGLQPRQ VAIWFQNRRA 180 RWKTKQLEKD YDALKRQLDA VKAENDALLN HNKKLQAEIV ALKGREAASE LINLNKETEA 240 SCSNRSENSS EINLDISRTP PPDAAALDAA PTAHHHHHGG GGGGGGMIPF YTSIARPASG 300 GGVDIDQLLH SSSGGAGGPK MEHHGGGGNV QAASVDTASF GNLLCGVDEP PPFWPWPDHQ 360 HFH |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 177 | 185 | RRARWKTKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00225 | DAP | Transfer from AT1G69780 | Download |
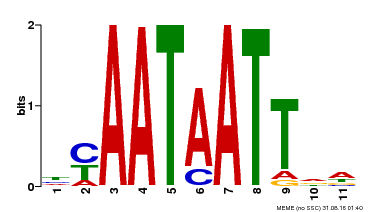
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC073556 | 0.0 | AC073556.11 Oryza sativa chromosome 3 BAC OSJNBa0091P11 genomic sequence, complete sequence. | |||
| GenBank | AC144491 | 0.0 | AC144491.1 Oryza sativa Japonica Group chromosome 3 clone OSJNAa0091P11, complete sequence. | |||
| GenBank | AP014959 | 0.0 | AP014959.1 Oryza sativa Japonica Group DNA, chromosome 3, cultivar: Nipponbare, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015629266.1 | 0.0 | homeobox-leucine zipper protein HOX21 | ||||
| Swissprot | Q8S7W9 | 0.0 | HOX21_ORYSJ; Homeobox-leucine zipper protein HOX21 | ||||
| TrEMBL | A0A0D3FED9 | 0.0 | A0A0D3FED9_9ORYZ; Uncharacterized protein | ||||
| STRING | OBART03G05300.1 | 0.0 | (Oryza barthii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2289 | 38 | 94 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G69780.1 | 1e-70 | HD-ZIP family protein | ||||



