 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OBART03G06570.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1007aa MW: 112741 Da PI: 5.8267 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 141.8 | 2e-44 | 20 | 116 | 3 | 98 |
CG-1 3 ke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfq 95
ke ++rwl+++ei++iL+n+++++++ e+++rp+sgsl+L++rk++ryfrkDG++w+kk+dgktv+E+he+LK g+++vl+cyYah+een +fq
OBART03G06570.1 20 KEaQRRWLRPTEICEILKNYRSFRIAPEPPNRPPSGSLFLFDRKVLRYFRKDGHNWRKKRDGKTVKEAHERLKSGSIDVLHCYYAHGEENINFQ 113
55599***************************************************************************************** PP
CG-1 96 rrc 98
rr
OBART03G06570.1 114 RRR 116
*96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 64.466 | 15 | 142 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 2.0E-49 | 18 | 126 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 5.5E-38 | 21 | 116 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 5.0E-7 | 418 | 503 | IPR013783 | Immunoglobulin-like fold |
| SuperFamily | SSF81296 | 1.52E-17 | 420 | 505 | IPR014756 | Immunoglobulin E-set |
| CDD | cd00102 | 0.00336 | 420 | 507 | No hit | No description |
| Pfam | PF01833 | 5.4E-8 | 420 | 495 | IPR002909 | IPT domain |
| SuperFamily | SSF48403 | 9.48E-17 | 598 | 712 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 4.3E-18 | 611 | 717 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 2.05E-13 | 612 | 710 | No hit | No description |
| PROSITE profile | PS50297 | 18.059 | 618 | 722 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 8.763 | 618 | 650 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 8.843 | 651 | 683 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 0.013 | 651 | 680 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 1400 | 690 | 719 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 4.78E-8 | 822 | 875 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 0.075 | 824 | 846 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.407 | 825 | 854 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0033 | 826 | 845 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0022 | 847 | 869 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.578 | 848 | 872 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 2.0E-4 | 850 | 869 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1007 aa Download sequence Send to blast |
MAEGRRYAIA PQLDIEQILK EAQRRWLRPT EICEILKNYR SFRIAPEPPN RPPSGSLFLF 60 DRKVLRYFRK DGHNWRKKRD GKTVKEAHER LKSGSIDVLH CYYAHGEENI NFQRRRITCT 120 LYLYIIWKSS TGHDDVLQAS HADSPLSQLP SQTTEGESSV SGQASEYDET ESAYLKQNSV 180 QQIFIREEPD TTLSLGCGSM KMEVDLYQGL QATAPNTGFY SHGQDNLPMV LNESDLGTAF 240 NGPNSQIDLS LWIEAMKPDK GTHQIPLYQA PVPSEQSPFT GGPGIESFTF DEVYNNGLSI 300 KDVDGDDTDG ETPWQIPNAS GTFATVDSFQ QNDKTLEEAI NYPLLKTQSS SLSDIIKDGF 360 KKNDSFTRWM SKELAEVDDS QITSSSGVYW NSEEADNIIE ASSSDQYTLG PVLAQDQLFT 420 IVDFSPTWTY AGSKTRVFIK GNFLSSDEVK RLKWSCMFGE FEVPAEIIAD DTLVCHSPSH 480 KPGRVPFYVT CSNRLACSEV REFDFRPQYM DAPSPLGSTN KIYLQKRLDK LLSVEQDEIH 540 TTLSNPTKEI IDLSKKISSL MMNNDDWSEL LKLADDNEPA TDDKQDQFLQ NRIKEKLHIW 600 LLHKVGDGGK GPSVLDEEGQ GVLHLAAALG YDWAIRPTIA AGVNINFRDA HGWTALHWAA 660 FCGRERTVVA LIALGAAPGA VTDPTPSFPS GSTPADLASA NGHKGISGFL AESSLTSHLQ 720 TLNLKEAMRS SAGEISGLPG IVNVADRSAS PLAVEGHQTG SMGDSLGAVR NAAQAAARIY 780 QVFRMQSFQR KQAVQYEDEN GAISDERAMS LLSAKPSKPA QLDPLHAAAT RIQNKFRGWK 840 GRKEFLLIRQ RIVKIQAHVR GHQVRKHYRK IIWSVGIVEK VILRWRRRGA GLRGFRPTEN 900 AVTESTSSSS GNVTQNRPAE NDYDFLQEGR KQTEERLQKA LARVKSMVQY PDARDQYQRI 960 LTVVTKMQES QAMQEKMLEE STEMDEGLLM SEFKELWDDD MPTPGYF |
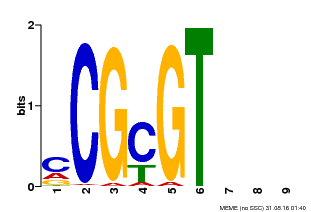
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00042 | PBM | Transfer from AT2G22300 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK066651 | 0.0 | AK066651.1 Oryza sativa Japonica Group cDNA clone:J013074G10, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015631248.1 | 0.0 | calmodulin-binding transcription activator 1 isoform X1 | ||||
| TrEMBL | A0A0D3FEU7 | 0.0 | A0A0D3FEU7_9ORYZ; Uncharacterized protein | ||||
| STRING | OBART03G06570.2 | 0.0 | (Oryza barthii) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G22300.2 | 0.0 | signal responsive 1 | ||||



