 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OBART07G08110.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 515aa MW: 56007.9 Da PI: 8.9755 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 54.1 | 3.7e-17 | 200 | 249 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV++++++grW+++I+d + k+++lg f+ta Aa+a+++a+ k++g
OBART07G08110.2 200 SQYRGVTFYRRTGRWESHIWD------CgKQVYLGGFDTAHAAARAYDRAAIKFRG 249
78*******************......55************************998 PP
| |||||||
| 2 | AP2 | 42.1 | 2.1e-13 | 292 | 342 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpseng...krkrfslgkfgtaeeAakaaiaarkkleg 55
s+y+GV+ +k grW+A+ g +k+++lg f+++ eAa+a+++a+ +++g
OBART07G08110.2 292 SKYRGVTLHK-CGRWEAR---M---GqllGKKYIYLGLFDSEIEAARAYDRAAIRFNG 342
89********.7******...5...334336**********99**********99987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 2.4E-9 | 200 | 249 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 6.47E-16 | 200 | 258 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 3.1E-31 | 201 | 263 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 17.82 | 201 | 257 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 1.02E-10 | 201 | 256 | No hit | No description |
| Gene3D | G3DSA:3.30.730.10 | 5.9E-17 | 201 | 257 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 5.0E-9 | 292 | 342 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 3.34E-26 | 292 | 352 | No hit | No description |
| SuperFamily | SSF54171 | 8.5E-17 | 292 | 351 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 4.2E-33 | 293 | 356 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 7.2E-16 | 293 | 350 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 16.107 | 293 | 350 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.4E-6 | 294 | 305 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 2.4E-6 | 332 | 352 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 515 aa Download sequence Send to blast |
MVLPAIPFVS LTSHPSLPPF SLLLHLHCHG SLRRSSLPRG RRRRLPPLGE RDGGAVGATQ 60 LALSPRGWWG CARGERLVSM VLDLNVESPG GSAATSSSST PPPPPDGGGG GYFRFDLLGG 120 SPDEDGCSPP VMTRQLFPSP SAVVALAGDG SSTPPPTMPT PAAAGEGPWP RRPADLGVAQ 180 SQRSPAGGKK SRRGPRSRSS QYRGVTFYRR TGRWESHIWD CGKQVYLGGF DTAHAAARAY 240 DRAAIKFRGL DADINFNLND YEDDLKQMRN WTKEEFVHIL RRQSTGFARG SSKYRGVTLH 300 KCGRWEARMG QLLGKKYIYL GLFDSEIEAA RAYDRAAIRF NGREAVTNFD PSSYDGDVLP 360 ETDNEVVDGD IIDLNLRISQ PNVHELKSDG TLTGFQLNCD SPEASSSVVT QPISPQWPVL 420 PQGTSMSQHP HLYASPCPGF FVNLREVPME KRPELGPQSF PTSWSWQMQG SPLPLLPTAA 480 SSGFSTGTVA DAARAPSSRP HPFPGHHQFY FPPTA |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor (By similarity). Involved in spikelet transition. Together with IDS1, controls synergistically inflorescence architecture and floral meristem establishment via the regulation of spatio-temporal expression of B- and E-function floral organ identity genes in the lodicules and of spikelet meristem genes. Prevents lemma and palea elongation as well as grain growth. Regulates the transition from spikelet meristem to floral meristem, spikelet meristem determinancy and the floral organ development (By similarity). {ECO:0000250|UniProtKB:P47927, ECO:0000250|UniProtKB:Q8H443}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00615 | PBM | Transfer from AT2G28550 | Download |
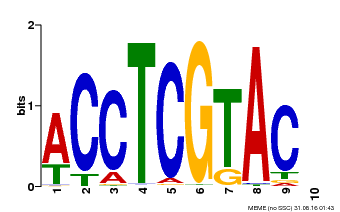
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | CT831975 | 0.0 | CT831975.1 Oryza sativa (indica cultivar-group) cDNA clone:OSIGCRA106E13, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015647891.1 | 0.0 | ethylene-responsive transcription factor RAP2-7 isoform X1 | ||||
| Swissprot | B8B8J2 | 0.0 | AP23_ORYSI; APETALA2-like protein 3 | ||||
| TrEMBL | A0A0D3GNY3 | 0.0 | A0A0D3GNY3_9ORYZ; Uncharacterized protein | ||||
| STRING | OBART07G08110.1 | 0.0 | (Oryza barthii) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G28550.3 | 1e-92 | related to AP2.7 | ||||



