 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OBART07G20460.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 116aa MW: 13369.4 Da PI: 10.0253 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 55.7 | 1.1e-17 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd +lv +++++G+g+W++++ g+ R+ k+c++rw +yl
OBART07G20460.1 14 KGPWTPEEDIILVSYIQEHGPGNWRSVPINTGLMRCSKSCRLRWTNYL 61
79********************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 49 | 1.4e-15 | 67 | 112 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg++T+ E+ ++v++ +lG++ W++Ia++++ Rt++++k++w+++l
OBART07G20460.1 67 RGNFTAHEEGIIVHLQSLLGNR-WAAIASYLP-QRTDNDIKNYWNTHL 112
89********************.*********.************996 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 5.9E-25 | 5 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 25.517 | 9 | 65 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 5.62E-31 | 11 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 4.4E-13 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.9E-16 | 14 | 61 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.03E-11 | 16 | 61 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 8.3E-24 | 65 | 115 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 4.1E-13 | 66 | 114 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 17.427 | 66 | 116 | IPR017930 | Myb domain |
| Pfam | PF00249 | 2.7E-13 | 67 | 112 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.08E-9 | 69 | 112 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009414 | Biological Process | response to water deprivation | ||||
| GO:0009416 | Biological Process | response to light stimulus | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0010118 | Biological Process | stomatal movement | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 116 aa Download sequence Send to blast |
MGRPPCCDKE GIKKGPWTPE EDIILVSYIQ EHGPGNWRSV PINTGLMRCS KSCRLRWTNY 60 LRPGIKRGNF TAHEEGIIVH LQSLLGNRWA AIASYLPQRT DNDIKNYWNT HLKKKP |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h8a_C | 2e-26 | 12 | 115 | 25 | 127 | MYB TRANSFORMING PROTEIN |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in the regulation of gene (e.g. drought-regulated and flavonoid biosynthetic genes) expression and stomatal movements leading to negative regulation of responses to drought and responses to other physiological stimuli (e.g. light). {ECO:0000269|PubMed:22018045}. | |||||
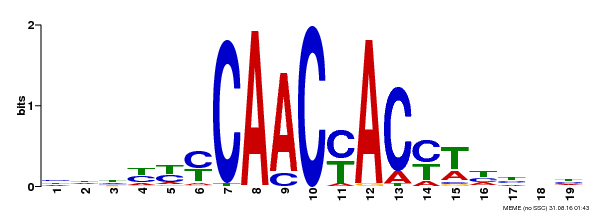
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00132 | DAP | Transfer from AT1G08810 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by abscisic acid (ABA) and osmotic stress (salt stress). {ECO:0000269|PubMed:22018045}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK241549 | 0.0 | AK241549.1 Oryza sativa Japonica Group cDNA, clone: J065176M15, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015616542.1 | 1e-82 | myb-related protein 306 | ||||
| Refseq | XP_015619015.1 | 1e-82 | myb-related protein 306 | ||||
| Refseq | XP_015698193.1 | 4e-83 | PREDICTED: myb-related protein 306-like | ||||
| Swissprot | B3VTV7 | 7e-79 | MYB60_VITVI; Transcription factor MYB60 | ||||
| TrEMBL | A0A0E0MBY8 | 2e-82 | A0A0E0MBY8_ORYPU; Uncharacterized protein | ||||
| STRING | OPUNC11G01450.1 | 3e-83 | (Oryza punctata) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP229 | 38 | 296 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G08810.1 | 9e-81 | myb domain protein 60 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



