 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OGLUM01G09290.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 993aa MW: 108649 Da PI: 4.7908 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 58.4 | 1.6e-18 | 51 | 97 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd +l +av+ + g++Wk+Ia++++ Rt+ qc +rwqk+l
OGLUM01G09290.1 51 KGNWTPEEDAILSRAVQTYNGKNWKKIAECFP-DRTDVQCLHRWQKVL 97
79******************************.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 62.3 | 9.6e-20 | 103 | 149 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+W++eEde++v+ v++lG++ W+tIa+ ++ gR +kqc++rw+++l
OGLUM01G09290.1 103 KGPWSKEEDEIIVQMVNKLGPKKWSTIAQALP-GRIGKQCRERWYNHL 149
79******************************.*************97 PP
| |||||||
| 3 | Myb_DNA-binding | 48.5 | 2e-15 | 155 | 197 | 1 | 45 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwq 45
+ +WT+eE+ l++a++++G++ W+ + ++ gRt++++k++w+
OGLUM01G09290.1 155 KEAWTQEEEITLIHAHRMYGNK-WAELTKFLP-GRTDNSIKNHWN 197
579*******************.*********.***********8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 16.383 | 46 | 97 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 3.31E-16 | 47 | 103 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 5.1E-17 | 50 | 99 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.3E-16 | 51 | 97 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.0E-24 | 53 | 114 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.26E-14 | 53 | 97 | No hit | No description |
| PROSITE profile | PS51294 | 31.183 | 98 | 153 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 9.35E-31 | 100 | 196 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 3.2E-18 | 102 | 151 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.2E-17 | 103 | 149 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 6.02E-16 | 105 | 149 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 1.8E-25 | 115 | 156 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 19.815 | 154 | 204 | IPR017930 | Myb domain |
| SMART | SM00717 | 2.2E-14 | 154 | 202 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 6.5E-14 | 155 | 198 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.1E-21 | 157 | 204 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 4.15E-11 | 157 | 197 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0032875 | Biological Process | regulation of DNA endoreduplication | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003713 | Molecular Function | transcription coactivator activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 993 aa Download sequence Send to blast |
MMTSDNGKAP DKDGEPSGPP SAPQEGEISN EPKRRRPLNG RTTGPTRRST KGNWTPEEDA 60 ILSRAVQTYN GKNWKKIAEC FPDRTDVQCL HRWQKVLNPE LVKGPWSKEE DEIIVQMVNK 120 LGPKKWSTIA QALPGRIGKQ CRERWYNHLN PGINKEAWTQ EEEITLIHAH RMYGNKWAEL 180 TKFLPGRTDN SIKNHWNSSV KKKVNSYMSS GLLTQVSCLP LNEYSANCNS SPAMTQQNSE 240 DSGCFAVREV ENSSGCSQSS LAKVSCSQVH DTTVPLGCDL QVNANFDKNE AHDSQSSMGP 300 QACYTSAEAV ASALPAVHCH VSSSNLDPDQ HLQEDFAQGL NLDMTIDEMP TVPSFADNQT 360 VCSIENHERS LEPYDVAMEV PLSMLSSDSG AEQKLHFMSE ADFNSPNCLK SELWQDISLQ 420 GLLSGPDAVE ADSFSRSNHQ SDVYSSEADT HFLAPPYMPQ TSNSSSVMGL ADDQSPQMSV 480 PPSLICSNAM TDDAPFDNRP GRKEMPLSQA EVVTQSSSSS GDAEMFANPG CSNDRHVPSS 540 TMESIPECGD QQVTNAEEPE ASLEKEPSLT QSVTAPDEQD KGALFYEPPR FPSLDVPFVS 600 CDLVTSGDLQ EFSPLGIRQL MHSTMNVCTP MRLWGSPTHD ESTGVLLKSA AKSFICTPSI 660 LKKRHRDLLS PIPDKRIEKK YGTEKDRGVS DTSSTGIQTS CINATKDDAL ITTVLRIERS 720 ASSKSLEKKL VFSDENKENL GYTTEQTKDG QSAGNDEHMD EQTTGERSSA TNVATNDDLS 780 GNLQPAGILI EHSGDDPISP DYGKNTMNQK LNTNVKSLSV CKEGVCAKSK PTELIVEKSS 840 PCINVDYEYV NILADTPGIK RGLESPSAWK SPWFVDMHFQ GSYFTSPADS YDALGLMKQI 900 NVQTAAALVE AREVLASGGQ CDNISSDKEN TGNPDAKKEP GTTKLQTKIM AEGRVLDFEC 960 TTPVRSSDKN SGSNLGRYLS SPIPSSHLLK SFR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h88_C | 1e-65 | 51 | 204 | 6 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 1e-65 | 51 | 204 | 6 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00505 | DAP | Transfer from AT5G11510 | Download |
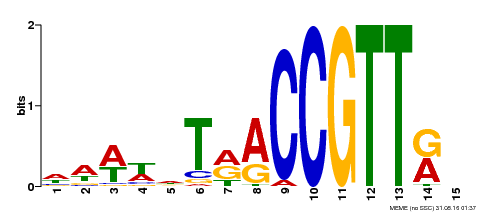
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK318601 | 0.0 | AK318601.1 Oryza sativa Japonica Group cDNA, clone: J100066E24, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015625869.1 | 0.0 | transcription factor MYB3R-1 | ||||
| TrEMBL | A0A0D9Y5K1 | 0.0 | A0A0D9Y5K1_9ORYZ; Uncharacterized protein | ||||
| STRING | OGLUM01G09290.1 | 0.0 | (Oryza glumipatula) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2495 | 38 | 79 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G11510.1 | 1e-136 | myb domain protein 3r-4 | ||||



