 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OGLUM01G09830.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 785aa MW: 84938.6 Da PI: 10.0753 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 38.9 | 1.6e-12 | 631 | 677 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCC.C...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPk.askapskKlsKaeiLekAveYIksLq 55
+h e+Er+RR+++N++f Lr ++P+ + K++Ka+ L A+ YI +Lq
OGLUM01G09830.1 631 NHVEAERQRREKLNQRFYALRAVVPNiS------KMDKASLLGDAITYITDLQ 677
799***********************66......*****************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 9.2E-50 | 228 | 413 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 16.831 | 627 | 676 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.57E-18 | 630 | 693 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 7.31E-15 | 630 | 681 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 8.9E-18 | 631 | 690 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 5.1E-10 | 631 | 677 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.9E-16 | 633 | 682 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 785 aa Download sequence Send to blast |
MNKPPARKAE KSPWVGDRAG SPAPSEPRTG SARLGSGSNP TGVEKAKAPS FSFHTSPPLF 60 FSLSTSQVAS LLRPRNPLTN QRNLRHHHHH HHRLLRARRI RGTLALAWGV GGGRSGRRRR 120 RGGGCSDSAS GGWGGPPLSS WGGAEETPPV LDSVAAAPIR AREPQHGIMV MKMEADEDGA 180 NGGTGGTWTD EDRALTASVL GTDAFAYLTK GGGAISEGLV AASLPVDLQN RLQELVESDR 240 PGAGWNYAIF WQLSRTKSGD LVLGWGDGSC REPRDGEMGP AASAGSDEAK QRMRKRVLQR 300 LHSAFGGVDE EDYAPGIDQV TDTEMFFLAS MYFAFPRRAG GPGQVFAAGV PLWIPNTERN 360 VFPANYCYRG YLANAAGFRT IVLVPFETGV LELGSMQQVA ESSDTLQTIR SVFAGAIGNK 420 AGVQRHEGSG PTDKSPGLAK IFGKDLNLGR PSAGPGTGMS KADERSWEQR TGGGSSLLPN 480 VQRGLQNFTW SQARGLNSHQ QKFGNGILIV SNEATPRNNG VVDSSTATQF QLQKAPPLQK 540 LPQLQKSHQL VKPQQLVSQQ QLQPQAPRQI DFSAGTSSKP GVLTKKPAGI DGDGAEVDGL 600 CKDEGPPPAL EDRRPRKRGR KPANGREEPL NHVEAERQRR EKLNQRFYAL RAVVPNISKM 660 DKASLLGDAI TYITDLQKKL KEMEVERERL IESGMIDPRD RTPRPEVDIQ VVQDEVLVRV 720 MSPMESHPVR AIFQAFEEAE VHAGESKITS NNGTAVHSFI IKCPGAEQQT REKVIAAMSR 780 VMNSG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5gnj_A | 2e-26 | 625 | 687 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_B | 2e-26 | 625 | 687 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_E | 2e-26 | 625 | 687 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_F | 2e-26 | 625 | 687 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_G | 2e-26 | 625 | 687 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_I | 2e-26 | 625 | 687 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_M | 2e-26 | 625 | 687 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_N | 2e-26 | 625 | 687 | 2 | 64 | Transcription factor MYC2 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 612 | 620 | RRPRKRGRK |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00100 | PBM | Transfer from AT1G01260 | Download |
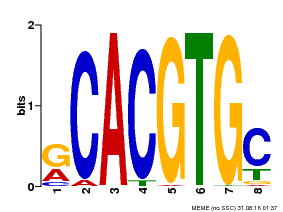
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | CT833046 | 0.0 | CT833046.1 Oryza sativa (indica cultivar-group) cDNA clone:OSIGCRA221E23, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015621308.1 | 0.0 | transcription factor bHLH13 | ||||
| TrEMBL | A0A0D9Y5R9 | 0.0 | A0A0D9Y5R9_9ORYZ; Uncharacterized protein | ||||
| STRING | OGLUM01G09830.1 | 0.0 | (Oryza glumipatula) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP5174 | 38 | 60 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G01260.3 | 1e-70 | bHLH family protein | ||||



