 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OGLUM01G13370.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 650aa MW: 68130.2 Da PI: 6.6739 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 49.4 | 8.3e-16 | 401 | 447 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K +Ka++L +A+eY+k Lq
OGLUM01G13370.1 401 VHNLSERRRRDRINEKMRALQELIPNC-----NKIDKASMLDEAIEYLKTLQ 447
5*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 2.96E-17 | 394 | 451 | No hit | No description |
| SuperFamily | SSF47459 | 1.96E-20 | 395 | 456 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.981 | 397 | 446 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 2.4E-20 | 401 | 455 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 2.8E-13 | 401 | 447 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 2.1E-17 | 403 | 452 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 650 aa Download sequence Send to blast |
MHVFVFGSVF CVCRSDGNDF AELLWENGQA VVHGRKKHPQ PAFPPFGFFG GTGGGGGGSS 60 SRAQEKQPGG IDAFAKVGGG FGALGMAPAV HDFASGFGAT TQDNGDDDTV PWIHYPIIDD 120 EDAAAPAALA AADYGSDFFS ELQAAAAAAA AAAPPTDLAS LPASNHNGAT NNRNAPVANT 180 TTREPSKESH GGLSVPTTRA EPQPQPQLAA AKLPRSSGSG GGEGVMNFSL FSRPAVLARA 240 TLESAQRTQG TDNKASNVTA SNRVESTVVQ TASGPRSAPA FADQRAAAWP PQPKEMPFAS 300 TAAAPMAPAV NLHHEMGRDR AGRTMPVHRT ETRKAPEATV ATSSVCSGNG AGSDELWRQQ 360 KRKCQAQAEC SASQDDDLDD EPGVLRKSGT RSTKRSRTAE VHNLSERRRR DRINEKMRAL 420 QELIPNCNKI DKASMLDEAI EYLKTLQLQV QMMSMGTGLC IPPMLLPTAM QHLQIPPMAH 480 FPHLGMGLGY GMGVFDMSNT GALQMPPMPG AHFPCPMIPG ASPQGLGIPG TSTMPMFGVP 540 GQTILSSASS VPPFASLAGL PVRPSGVPQV SGAMANMVQD QQQGIANQQQ QCLNKEAIQG 600 ANPGDSQMQI IMQGDNENFR IPSSAQTKSS QFSDGTGKGT NARERDGAET |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 405 | 410 | ERRRRD |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that may act as negative regulator of phyB-dependent light signal transduction. {ECO:0000269|PubMed:17485859}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
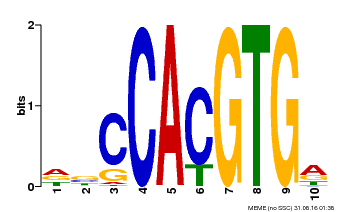
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated by light in dark-grown etiolated seedlings. {ECO:0000269|PubMed:17485859}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK102252 | 0.0 | AK102252.1 Oryza sativa Japonica Group cDNA clone:J033088H11, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015619034.1 | 0.0 | transcription factor PHYTOCHROME INTERACTING FACTOR-LIKE 15-like isoform X2 | ||||
| Swissprot | Q0JNI9 | 0.0 | PIL15_ORYSJ; Transcription factor PHYTOCHROME INTERACTING FACTOR-LIKE 15 | ||||
| TrEMBL | A0A0D9Y6Z4 | 0.0 | A0A0D9Y6Z4_9ORYZ; Uncharacterized protein | ||||
| STRING | OGLUM01G13370.1 | 0.0 | (Oryza glumipatula) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3527 | 37 | 75 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 3e-34 | phytochrome interacting factor 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



