 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OGLUM01G31260.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | RAV | ||||||||
| Protein Properties | Length: 396aa MW: 41534.6 Da PI: 9.1396 | ||||||||
| Description | RAV family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 37.6 | 5.3e-12 | 79 | 127 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
s++kGV + +grW A+I++ r +r++lg+f +++Aa+a++ a+++++g
OGLUM01G31260.1 79 SKFKGVVPQP-NGRWGAQIYE------RhQRVWLGTFAGEDDAARAYDVAAQRFRG 127
79****9888.8*********......44**********99*************98 PP
| |||||||
| 2 | B3 | 97.6 | 8e-31 | 215 | 307 | 1 | 86 |
EEEE-..-HHHHTT-EE--HHH.HTT.......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-S CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh.......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldg 86
f+k+ tpsdv+k++rlv+pk++ae+h g + +++ l +ed g++W+++++y+++s++yvltkGW++Fvk++gL +gD+v F+++
OGLUM01G31260.1 215 FDKTVTPSDVGKLNRLVIPKQHAEKHfplqlpsAGGESKGVLLNFEDDAGKVWRFRYSYWNSSQSYVLTKGWSRFVKEKGLHAGDVVGFYRSA 307
89*******************************5556799*************************************************9653 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF54171 | 4.25E-16 | 79 | 135 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 8.31E-23 | 79 | 135 | No hit | No description |
| Pfam | PF00847 | 1.4E-7 | 79 | 127 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 19.52 | 80 | 135 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 4.5E-19 | 80 | 135 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 3.0E-26 | 80 | 141 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:2.40.330.10 | 8.4E-38 | 211 | 309 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 2.06E-29 | 213 | 318 | No hit | No description |
| SuperFamily | SSF101936 | 5.62E-30 | 213 | 308 | IPR015300 | DNA-binding pseudobarrel domain |
| SMART | SM01019 | 2.2E-21 | 215 | 323 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 13.634 | 215 | 326 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 8.3E-28 | 215 | 307 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 396 aa Download sequence Send to blast |
MDSSSCLVDD TNSGGSSTDK LRALAAAAAE TAPLERMGSG ASAVVDAAEP GAEADSGSGG 60 RVCGGGGGAG GAGGKLPSSK FKGVVPQPNG RWGAQIYERH QRVWLGTFAG EDDAARAYDV 120 AAQRFRGRDA VTNFRPLAEA DPDAAAELRF LATRSKAEVV DMLRKHTYFD ELAQSKRTFA 180 ASTPSAATTT ASLSNGHLSS PRSPFAPAAA RDHLFDKTVT PSDVGKLNRL VIPKQHAEKH 240 FPLQLPSAGG ESKGVLLNFE DDAGKVWRFR YSYWNSSQSY VLTKGWSRFV KEKGLHAGDV 300 VGFYRSAASA GDDGKLFIDC KLVRSTGAAL ASPADQPAPS PVKAVRLFGV DLLTAPTPAP 360 APVEQMAGCK RARDLAATTP PQAAAFKKQC IELALV |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wid_A | 9e-49 | 214 | 321 | 13 | 115 | DNA-binding protein RAV1 |
| 5os9_A | 8e-49 | 209 | 319 | 2 | 109 | B3 domain-containing transcription factor NGA1 |
| 5os9_B | 8e-49 | 209 | 319 | 2 | 109 | B3 domain-containing transcription factor NGA1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 58 | 66 | GGRVCGGGG |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00025 | PBM | Transfer from AT1G68840 | Download |
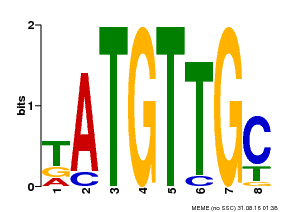
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK241984 | 0.0 | AK241984.1 Oryza sativa Japonica Group cDNA, clone: J075097P17, full insert sequence. | |||
| GenBank | AP003450 | 0.0 | AP003450.3 Oryza sativa Japonica Group genomic DNA, chromosome 1, PAC clone:P0034C09. | |||
| GenBank | AP014957 | 0.0 | AP014957.1 Oryza sativa Japonica Group DNA, chromosome 1, cultivar: Nipponbare, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015622542.1 | 0.0 | AP2/ERF and B3 domain-containing protein Os01g0693400 | ||||
| Swissprot | Q8RZX9 | 0.0 | Y1934_ORYSJ; AP2/ERF and B3 domain-containing protein Os01g0693400 | ||||
| TrEMBL | A0A0D9YDI3 | 0.0 | A0A0D9YDI3_9ORYZ; Uncharacterized protein | ||||
| STRING | OGLUM01G31260.1 | 0.0 | (Oryza glumipatula) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP456 | 38 | 203 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G68840.2 | 1e-105 | related to ABI3/VP1 2 | ||||



