 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OGLUM03G06680.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 961aa MW: 108109 Da PI: 6.0014 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 176.8 | 2.7e-55 | 20 | 136 | 3 | 118 |
CG-1 3 ke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfq 95
ke ++rwl+++ei++iL+n+++++++ e+++rp+sgsl+L++rk++ryfrkDG++w+kk+dgktv+E+he+LK g+++vl+cyYah+een +fq
OGLUM03G06680.2 20 KEaQRRWLRPTEICEILKNYRSFRIAPEPPNRPPSGSLFLFDRKVLRYFRKDGHNWRKKRDGKTVKEAHERLKSGSIDVLHCYYAHGEENINFQ 113
55599***************************************************************************************** PP
CG-1 96 rrcywlLeeelekivlvhylevk 118
rr+yw+Lee++ +ivlvhylevk
OGLUM03G06680.2 114 RRSYWMLEEDYMHIVLVHYLEVK 136
********************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 80.6 | 15 | 141 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 5.2E-76 | 18 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 1.4E-48 | 21 | 135 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 4.7E-7 | 372 | 457 | IPR013783 | Immunoglobulin-like fold |
| Pfam | PF01833 | 5.1E-8 | 374 | 449 | IPR002909 | IPT domain |
| SuperFamily | SSF81296 | 1.43E-17 | 374 | 459 | IPR014756 | Immunoglobulin E-set |
| CDD | cd00102 | 0.00319 | 374 | 461 | No hit | No description |
| SuperFamily | SSF48403 | 8.86E-17 | 552 | 666 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 4.2E-18 | 565 | 671 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.92E-13 | 566 | 664 | No hit | No description |
| PROSITE profile | PS50297 | 18.059 | 572 | 676 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 8.763 | 572 | 604 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 0.013 | 605 | 634 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 8.843 | 605 | 637 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 1400 | 644 | 673 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 4.51E-8 | 776 | 829 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 0.075 | 778 | 800 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.407 | 779 | 808 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0031 | 780 | 799 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0022 | 801 | 823 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.578 | 802 | 826 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 1.9E-4 | 804 | 823 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0071275 | Biological Process | cellular response to aluminum ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 961 aa Download sequence Send to blast |
MAEGRRYAIA PQLDIEQILK EAQRRWLRPT EICEILKNYR SFRIAPEPPN RPPSGSLFLF 60 DRKVLRYFRK DGHNWRKKRD GKTVKEAHER LKSGSIDVLH CYYAHGEENI NFQRRSYWML 120 EEDYMHIVLV HYLEVKIFIR EEPDTTLSLG CGSMKMEVDL YQGLQATAPN TGFYSHGQDN 180 LPVVLNESDL GTAFNGPNSQ FDLSLWIEAM KPDKGTHQIP LYQAPVPSEQ SPFTGGPGIE 240 SFTFDEVYNN GLSIRDVDGD DTDGETPWQI PNASGTFATV DSFQQNDKTL EEAINYPLLK 300 TQSSSLSDII KDSFKKNDSF TRWMSKELAE VDDSQITSSS GVYWNSEEAD NIIEASSSDQ 360 YTLGPVLAQD QLFTIVDFSP TWTYAGSKTR VFIKGNFLSS DEVKRLKWSC MFGEFEVPAE 420 IIADDTLVCH SPSHKPGRVP FYVTCSNRLA CSEVREFDFR PQYMDAPSPL GSTNKIYLQK 480 RLDKLLSVEQ DEIQTTLSNP TKEIIDLSKK ISSLMMNNDD WSELLKLADD NEPATDDKQD 540 QFLQNRIKEK LHIWLLHKVG DGGKGPSVLD EEGQGVLHLA AALGYDWAIR PTIAAGVNIN 600 FRDAHGWTAL HWAAFCGRER TVVALIALGA APGAVTDPTP SFPSGSTPAD LASANGHKGI 660 SGFLAESSLT SHLQTLNLKE AMRSSAGEIS GLPGIVNVAD RSASPLAVEG HQTGSMGDSL 720 GAVRNAAQAA ARIYQVFRMQ SFQRKQAVQY EDENGAISDE RAMSLLSAKP SKPAQLDPLH 780 AAATRIQNKF RGWKGRKEFL LIRQRIVKIQ AHVRGHQVRK HYRKIIWSVG IVEKVILRWR 840 RRGAGLRGFR PTENAVTEST SSSSGNVTQN RPAENDYDFL QEGRKQTEER LQKALARVKS 900 MVQYPDARDQ YQRILTVVTK MQESQAMQEK MLEESTEMDE GLLMSEFKEL WDDDMPTPGY 960 F |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3'. Binds calmodulin in a calcium-dependent manner in vitro (PubMed:12218065). Regulates transcriptional activity in response to calcium signals (Probable). Involved in freezing tolerance in association with CAMTA1 and CAMTA2 (PubMed:23581962). Required for the cold-induced expression of DREB1B/CBF1, DREB1C/CBF2, ZAT12 and GOLS3 (PubMed:19270186). Involved in response to cold. Contributes together with CAMTA5 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). Involved together with CAMTA2 and CAMTA4 in the positive regulation of a general stress response (GSR) (PubMed:25039701). Involved in the regulation of GSR amplitude downstream of MEKK1 (PubMed:25157030). Involved in the regulation of a set of genes involved in defense responses against pathogens (PubMed:18298954). Involved in the regulation of both basal resistance and systemic acquired resistance (SAR) (PubMed:21900483). Acts as negative regulator of plant immunity (PubMed:19122675, PubMed:21900483, PubMed:22345509, PubMed:28407487). Binds to the promoter of the defense-related gene EDS1 and represses its expression (PubMed:19122675). Binds to the promoter of the defense-related gene NDR1 and represses its expression (PubMed:22345509). Involved in defense against insects (PubMed:23072934, PubMed:22371088). Required for tolerance to the generalist herbivore Trichoplusia ni, and contributes to the positive regulation of genes associated with glucosinolate metabolism (PubMed:23072934). Required for tolerance to Bradysia impatiens larvae. Mediates herbivore-induced wound response (PubMed:22371088). Required for wound-induced jasmonate accumulation (PubMed:23072934, PubMed:22371088). Involved in the regulation of ethylene-induced senescence by binding to the promoter of the senescence-inducer gene EIN3 and repressing its expression (PubMed:22345509). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:18298954, ECO:0000269|PubMed:19122675, ECO:0000269|PubMed:19270186, ECO:0000269|PubMed:21900483, ECO:0000269|PubMed:22345509, ECO:0000269|PubMed:22371088, ECO:0000269|PubMed:23072934, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25157030, ECO:0000269|PubMed:28351986, ECO:0000269|PubMed:28407487, ECO:0000305|PubMed:11925432}. | |||||
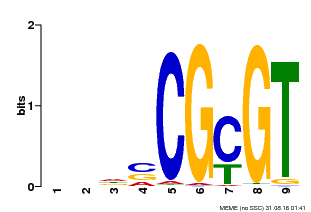
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00043 | PBM | Transfer from AT5G64220 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, salt, wounding, ethylene and methyl jasmonate (PubMed:11162426, PubMed:12218065). Induced by infection with the fungal pathogen Golovinomyces cichoracearum (powdery mildew) and the bacterial pathogen Pseudomonas syringae pv tomato strain DC3000 (PubMed:22345509). {ECO:0000269|PubMed:11162426, ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:22345509}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK066651 | 0.0 | AK066651.1 Oryza sativa Japonica Group cDNA clone:J013074G10, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015631250.1 | 0.0 | calmodulin-binding transcription activator 2 isoform X3 | ||||
| Swissprot | Q8GSA7 | 0.0 | CMTA3_ARATH; Calmodulin-binding transcription activator 3 | ||||
| TrEMBL | A0A0D9Z394 | 0.0 | A0A0D9Z394_9ORYZ; Uncharacterized protein | ||||
| STRING | OGLUM03G06680.1 | 0.0 | (Oryza glumipatula) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G22300.2 | 0.0 | signal responsive 1 | ||||



