 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OGLUM04G14880.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | MIKC_MADS | ||||||||
| Protein Properties | Length: 238aa MW: 27473.1 Da PI: 9.2889 | ||||||||
| Description | MIKC_MADS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SRF-TF | 86.5 | 1.5e-27 | 10 | 59 | 2 | 51 |
---SHHHHHHHHHHHHHHHHHHHHHHHHHHT-EEEEEEE-TTSEEEEEE- CS
SRF-TF 2 rienksnrqvtfskRrngilKKAeELSvLCdaevaviifsstgklyeyss 51
ri+n++ rqvtfskRrngi+KKA+EL +LCdaev ++ifsst++lyey+s
OGLUM04G14880.1 10 RIDNSTSRQVTFSKRRNGIFKKAKELAILCDAEVGLVIFSSTDRLYEYAS 59
8***********************************************86 PP
| |||||||
| 2 | K-box | 91 | 2.2e-30 | 75 | 170 | 3 | 98 |
K-box 3 kssgksleeakaeslqqelakLkkeienLqreqRhllGedLesLslkeLqqLeqqLekslkkiRskKnellleqieelqkkekelqeenkaLrk 96
+++++++ +++ + +q+e+a L+++++nLq+++R+l+G+dL+ L +keLq Le+qLe s++ iR+kK++l++++i+el +k +++en++L +
OGLUM04G14880.1 75 EQQHVANPNSELKFWQREAASLRQQLHNLQENHRQLMGQDLSGLGVKELQTLENQLEMSIRCIRTKKDQLMIDEIHELNRKGSLIHQENMELYR 168
56667788999**********************************************************************************9 PP
K-box 97 kl 98
k+
OGLUM04G14880.1 169 KV 170
97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00432 | 1.2E-38 | 1 | 60 | IPR002100 | Transcription factor, MADS-box |
| PROSITE profile | PS50066 | 31.16 | 1 | 61 | IPR002100 | Transcription factor, MADS-box |
| CDD | cd00265 | 7.30E-43 | 2 | 77 | No hit | No description |
| SuperFamily | SSF55455 | 1.01E-32 | 2 | 91 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 4.6E-27 | 3 | 23 | IPR002100 | Transcription factor, MADS-box |
| PROSITE pattern | PS00350 | 0 | 3 | 57 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF00319 | 7.2E-25 | 10 | 57 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 4.6E-27 | 23 | 38 | IPR002100 | Transcription factor, MADS-box |
| PRINTS | PR00404 | 4.6E-27 | 38 | 59 | IPR002100 | Transcription factor, MADS-box |
| Pfam | PF01486 | 6.1E-29 | 83 | 170 | IPR002487 | Transcription factor, K-box |
| PROSITE profile | PS51297 | 15.797 | 86 | 176 | IPR002487 | Transcription factor, K-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010440 | Biological Process | stomatal lineage progression | ||||
| GO:0048574 | Biological Process | long-day photoperiodism, flowering | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008134 | Molecular Function | transcription factor binding | ||||
| GO:0042803 | Molecular Function | protein homodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 238 aa Download sequence Send to blast |
MGRGKIVIRR IDNSTSRQVT FSKRRNGIFK KAKELAILCD AEVGLVIFSS TDRLYEYAST 60 SMKSVIDRYG RAKEEQQHVA NPNSELKFWQ REAASLRQQL HNLQENHRQL MGQDLSGLGV 120 KELQTLENQL EMSIRCIRTK KDQLMIDEIH ELNRKGSLIH QENMELYRKV NLIRQENAEL 180 YKKLYETGAE TEANRDSTTP YNFGVIEEAN TPARLELNPP SQQNDAEQTA PPKLGQKR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5f28_A | 4e-21 | 1 | 86 | 1 | 84 | MEF2C |
| 5f28_B | 4e-21 | 1 | 86 | 1 | 84 | MEF2C |
| 5f28_C | 4e-21 | 1 | 86 | 1 | 84 | MEF2C |
| 5f28_D | 4e-21 | 1 | 86 | 1 | 84 | MEF2C |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. | |||||
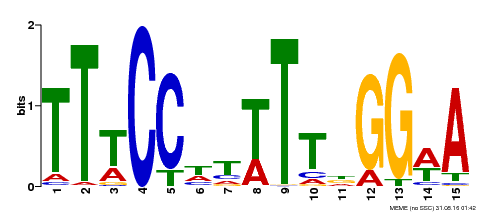
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00410 | DAP | Transfer from AT3G57230 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK243669 | 0.0 | AK243669.1 Oryza sativa Japonica Group cDNA, clone: J100089N11, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015636306.1 | 1e-171 | MADS-box transcription factor 27-like | ||||
| Swissprot | Q6EP49 | 1e-143 | MAD27_ORYSJ; MADS-box transcription factor 27 | ||||
| TrEMBL | A0A0D9ZLQ9 | 1e-176 | A0A0D9ZLQ9_9ORYZ; Uncharacterized protein | ||||
| STRING | OGLUM04G14880.1 | 1e-177 | (Oryza glumipatula) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP614 | 35 | 130 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G37940.1 | 3e-97 | AGAMOUS-like 21 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



