 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OGLUM06G25500.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 333aa MW: 35266.9 Da PI: 7.5813 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 40.2 | 7.7e-13 | 76 | 120 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r +WT+ E++++++a +++ + Wk+I +++g +t q++s+ qky
OGLUM06G25500.1 76 RESWTEPEHDKFLEALQLFDRD-WKKIEAYVG-SKTVIQIRSHAQKY 120
789*****************77.*********.*************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 3.36E-16 | 70 | 126 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 14.628 | 71 | 125 | IPR017930 | Myb domain |
| TIGRFAMs | TIGR01557 | 1.2E-18 | 74 | 123 | IPR006447 | Myb domain, plants |
| Gene3D | G3DSA:1.10.10.60 | 8.0E-6 | 75 | 116 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.7E-10 | 75 | 123 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.3E-10 | 76 | 120 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 8.35E-8 | 78 | 121 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 333 aa Download sequence Send to blast |
MSATPNPIDA SVVSAAAAVA GGGGGGDGGG GGGGKEKEKQ VVAAPLQPPM AVPAPAAAVG 60 EEARKVRKPY TITKSRESWT EPEHDKFLEA LQLFDRDWKK IEAYVGSKTV IQIRSHAQKY 120 FLKVQKNGTG EHLPPPRPKR KAAHPYPQKA SKNVSPAAIS QPPPLGEQGC VMSMDTSPVI 180 RNTNASAVVP SWDNSIAQPL SASRTQGTGA VATNNCSSSI ESPSTTWPTS EAVEQENMLR 240 PLRAMPDFAQ VYSFLGSIFD PDTSGHLQTL KAMDPIDVET VLLLMRNLSM NLTSPNFAAH 300 LSLLSSCNSG GDPIKSEGME NLGSPQSCHL PFM |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00625 | PBM | Transfer from PK02532.1 | Download |
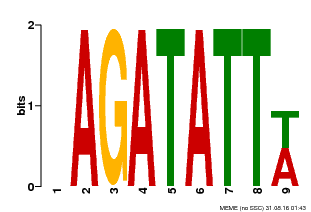
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK070871 | 0.0 | AK070871.1 Oryza sativa Japonica Group cDNA clone:J023073K02, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015643725.1 | 0.0 | protein REVEILLE 6 isoform X3 | ||||
| Swissprot | Q8H0W3 | 1e-95 | RVE6_ARATH; Protein REVEILLE 6 | ||||
| TrEMBL | B8B168 | 0.0 | B8B168_ORYSI; Uncharacterized protein | ||||
| STRING | ONIVA06G30670.1 | 0.0 | (Oryza nivara) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1096 | 38 | 111 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G52660.2 | 4e-83 | MYB_related family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



