 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OGLUM07G13800.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 930aa MW: 104276 Da PI: 7.4941 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 166.7 | 3.7e-52 | 31 | 146 | 3 | 117 |
CG-1 3 ke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfq 95
e +rw +++ei+aiL+n++++++++++ ++p sg+++Ly+rk+vr+frkDG++wkkkkdg+tv+E+hekLK+g+ e +++yYa++e++p+f
OGLUM07G13800.2 31 AEaAARWFRPNEIYAILANHARFKIHAQPVDKPVSGTVVLYDRKVVRNFRKDGHNWKKKKDGRTVQEAHEKLKIGNEERVHVYYARGEDDPNFF 124
45589***************************************************************************************** PP
CG-1 96 rrcywlLeeelekivlvhylev 117
rrcywlL+++le+ivlvhy+++
OGLUM07G13800.2 125 RRCYWLLDKDLERIVLVHYRQT 146
*******************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 74.858 | 26 | 152 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 2.9E-63 | 29 | 147 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 4.1E-45 | 33 | 145 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 4.2E-10 | 384 | 470 | IPR014756 | Immunoglobulin E-set |
| SuperFamily | SSF48403 | 3.11E-17 | 559 | 683 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 1.2E-17 | 569 | 683 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 5.39E-15 | 569 | 677 | No hit | No description |
| PROSITE profile | PS50297 | 17.953 | 585 | 689 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 9.2E-8 | 591 | 681 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 11.835 | 618 | 650 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 9.6E-6 | 618 | 647 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 580 | 657 | 686 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 8.5E-9 | 734 | 835 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| PROSITE profile | PS50096 | 6.54 | 769 | 795 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 1.5 | 784 | 806 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.785 | 785 | 812 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.002 | 787 | 805 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.016 | 807 | 829 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 10.274 | 808 | 832 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0092 | 810 | 829 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 2.4 | 890 | 912 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.084 | 892 | 920 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.044 | 893 | 912 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 930 aa Download sequence Send to blast |
MAGAGGWDPL VGSEIHGFLT YPDLNYEKLV AEAAARWFRP NEIYAILANH ARFKIHAQPV 60 DKPVSGTVVL YDRKVVRNFR KDGHNWKKKK DGRTVQEAHE KLKIGNEERV HVYYARGEDD 120 PNFFRRCYWL LDKDLERIVL VHYRQTAEEN AMAPPNPEPE VADVPTVNLI HYTSPLTSAD 180 STSGHTELSL PEEINSHGGI SASSETGNHD SSLEEFWANL LESSIKNDPK VVTSACGGIF 240 DKAMTIDITV VDQQINNGPK NSGNIFNTSM ASNAIPALNV VSETYATNHG LNQVNANHFG 300 ALKHQGDQTQ SLLASDVDSQ SDQFISSSVK SPMDGNTSIP NEVPARQNSL GLWKYLDDDS 360 PGLGDNPSSV PQSFCPVTNE RLLEINEISP EWAYSTETTK VVVIGNFYEQ YKHLAGSAMF 420 GVFGDQCVAG DIVQTGVYRF MVGPHTPGKV DFYLTLDGKT PISEICSFTY HVMHGSSLEA 480 RLPPSEDDYK RTNLQMQMRL ARLLFATNKK KIAPKLLVEG TKVANLMSAL SEKEWMDLWN 540 ILSDPEGTYV PVTESLLELV LRNRLQEWLV EMVMEGHKST GRDDLGQGAI HLCSFLGYTW 600 AIRLFSLSGF SLDFRDSSGW TALHWAAYHG RERMVATLLS AGANPSLVTD PTPESPAGLT 660 AADLAARQGY DGLAAYLAEK GLTAHFEAMS LSKDTEQSPS KTRLTKLQSE KFEHLSEQEL 720 CLKESLAAYR NAADAASNIQ AALRERTLKL QTKAIQLANP EIEASEIVAA MKIQHAFRNY 780 NRKKAMRAAA RIQSHFRTWK MRRNFINMRR QVIRIQAAYR GHQVRRQYRK VIWSVGIVEK 840 AILRWRKKRK GLRGIASGMP VVMTVDAEAE PASTAEEDFF QAGRQQAEDR FNRSVVRVQA 900 LFRSYKAQQE YRRMKIAHEE AKHDHLFIRL |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds calmodulin in a calcium-dependent manner in vitro. Binds to the DNA consensus sequence 5'-T[AC]CG[CT]GT[GT][GT][GT][GT]T[GT]CG-3'. {ECO:0000269|PubMed:16192280}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
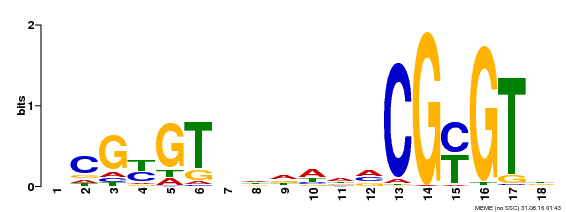
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK065480 | 0.0 | AK065480.1 Oryza sativa Japonica Group cDNA clone:J013025F20, full insert sequence. | |||
| GenBank | AK070976 | 0.0 | AK070976.1 Oryza sativa Japonica Group cDNA clone:J023073E17, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015646432.1 | 0.0 | calmodulin-binding transcription activator CBT-like isoform X2 | ||||
| Swissprot | Q7XHR2 | 0.0 | CBT_ORYSJ; Calmodulin-binding transcription activator CBT | ||||
| TrEMBL | A0A0E0AJU2 | 0.0 | A0A0E0AJU2_9ORYZ; Uncharacterized protein | ||||
| STRING | OGLUM07G13800.2 | 0.0 | (Oryza glumipatula) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP9831 | 29 | 33 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||



