 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OGLUM10G19510.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 699aa MW: 75103.8 Da PI: 6.6633 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 39.1 | 1.4e-12 | 524 | 569 | 4 | 54 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksL 54
+h e+Er+RR+++N++f Lr ++P+ K++Ka+ L A+ YI++L
OGLUM10G19510.1 524 NHVEAERQRREKLNQRFYALRAVVPNV-----SKMDKASLLGDAISYINEL 569
799***********************6.....5***************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 8.5E-51 | 68 | 248 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 17.39 | 520 | 569 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 2.69E-14 | 523 | 573 | No hit | No description |
| SuperFamily | SSF47459 | 2.49E-18 | 523 | 596 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 2.2E-18 | 524 | 591 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 4.6E-10 | 524 | 569 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 5.6E-16 | 526 | 575 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009269 | Biological Process | response to desiccation | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009867 | Biological Process | jasmonic acid mediated signaling pathway | ||||
| GO:0009963 | Biological Process | positive regulation of flavonoid biosynthetic process | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0043619 | Biological Process | regulation of transcription from RNA polymerase II promoter in response to oxidative stress | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0051090 | Biological Process | regulation of sequence-specific DNA binding transcription factor activity | ||||
| GO:2000068 | Biological Process | regulation of defense response to insect | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 699 aa Download sequence Send to blast |
MNLWTDDNAS MMEAFMASAD LPAFPWGAAS TPPPPPPPPH HHHQQQQQQV LPPPAAAPAA 60 AAFNQDTLQQ RLQSIIEGSR ETWTYAIFWQ SSIDVSTGAS LLGWGDGYYK GCDDDKRKQR 120 SSTPAAAAEQ EHRKRVLREL NSLIAGAGAA PDEAVEEEVT DTEWFFLVSM TQSFPNGLGL 180 PGQALFAAQP TWIATGLSSA PCDRARQAYT FGLRTMVCLP LATGVLELGS TDVIFQTGDS 240 IPRIRALFNL SAAAASSWPP HPDAASADPS VLWLADAPPM DMKDSISAAD ISVSKPPPPP 300 PHQIQHFENG STSTLTENPS PSVHAPTPSQ PAAPPQRQQQ QQQSSQAQQG PFRRELNFSD 360 FASNGGAAAP PFFKPETGEI LNFGNDSSSG RRNPSPAPPA ATASLTTAPG SLFSQHTPTL 420 TAAANDAKSN NQKRSMEATS RASNTNNHPA ATANEGMLSF SSAPTTRPST GTGAPAKSES 480 DHSDLEASVR EVESSRVVAP PPEAEKRPRK RGRKPANGRE EPLNHVEAER QRREKLNQRF 540 YALRAVVPNV SKMDKASLLG DAISYINELR GKLTALETDK ETLQSQMESL KKERDARPPA 600 PSGGGGDGGA RCHAVEIEAK ILGLEAMIRV QCHKRNHPAA RLMTALRELD LDVYHASVSV 660 VKDLMIQQVA VKMASRVYSQ DQLNAALYTR IAEPGTAAR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4rqw_A | 4e-62 | 63 | 251 | 4 | 195 | Transcription factor MYC3 |
| 4rqw_B | 4e-62 | 63 | 251 | 4 | 195 | Transcription factor MYC3 |
| 4rs9_A | 4e-62 | 63 | 251 | 4 | 195 | Transcription factor MYC3 |
| 4yz6_A | 4e-62 | 63 | 251 | 4 | 195 | Transcription factor MYC3 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 505 | 513 | KRPRKRGRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator involved in jasmonate (JA) signaling pathway during spikelet development. Binds to the G2 region G-box (5'-CACGTG-3') of the MADS1 promoter and thus directly regulates the expression of MADS1. Its function in MADS1 activation is abolished by TIFY3/JAZ1 which directly target MYC2 during spikelet development. {ECO:0000269|PubMed:24647160}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00084 | PBM | Transfer from AT1G32640 | Download |
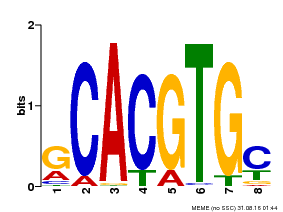
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC060755 | 0.0 | AC060755.9 Oryza sativa chromosome 10 BAC OSJNBa0003O19 genomic sequence, complete sequence. | |||
| GenBank | AK288082 | 0.0 | AK288082.1 Oryza sativa Japonica Group cDNA, clone: J075172G09, full insert sequence. | |||
| GenBank | AP014966 | 0.0 | AP014966.1 Oryza sativa Japonica Group DNA, chromosome 10, cultivar: Nipponbare, complete sequence. | |||
| GenBank | AY536428 | 0.0 | AY536428.1 Oryza sativa MYC protein mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015614012.2 | 0.0 | transcription factor MYC2-like | ||||
| Swissprot | Q336P5 | 0.0 | MYC2_ORYSJ; Transcription factor MYC2 | ||||
| TrEMBL | A0A0E0BE49 | 0.0 | A0A0E0BE49_9ORYZ; Uncharacterized protein | ||||
| STRING | OGLUM10G19510.1 | 0.0 | (Oryza glumipatula) | ||||
| STRING | OS10T0575000-01 | 0.0 | (Oryza sativa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP4452 | 31 | 57 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G32640.1 | 1e-131 | bHLH family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



