 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OGLUM12G09840.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1373aa MW: 152796 Da PI: 7.5874 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 12.8 | 0.00035 | 1254 | 1277 | 1 | 22 |
EEET..TTTEEESSHHHHHHHHHH CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt 22
++C C++sFs++ +L H r
OGLUM12G09840.1 1254 FPCDieGCDMSFSTQQDLLLHKRD 1277
789999***************985 PP
| |||||||
| 2 | zf-C2H2 | 11.9 | 0.00071 | 1337 | 1363 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH..T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt..H 23
y+C+ Cg++F+ s++ rH r+ H
OGLUM12G09840.1 1337 YECQepGCGQTFRFVSDFSRHKRKtgH 1363
899999****************99666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00545 | 1.9E-16 | 19 | 60 | IPR003349 | JmjN domain |
| PROSITE profile | PS51183 | 14.201 | 20 | 61 | IPR003349 | JmjN domain |
| Pfam | PF02375 | 3.4E-14 | 21 | 54 | IPR003349 | JmjN domain |
| PROSITE profile | PS51184 | 32.998 | 190 | 359 | IPR003347 | JmjC domain |
| SMART | SM00558 | 5.2E-43 | 190 | 359 | IPR003347 | JmjC domain |
| SuperFamily | SSF51197 | 9.61E-23 | 207 | 356 | No hit | No description |
| Pfam | PF02373 | 1.6E-34 | 223 | 342 | IPR003347 | JmjC domain |
| SMART | SM00355 | 9.7 | 1254 | 1276 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.907 | 1277 | 1306 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 15 | 1277 | 1301 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 1279 | 1301 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 1.58E-14 | 1288 | 1344 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 5.3E-11 | 1304 | 1331 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 8.6E-4 | 1307 | 1331 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.655 | 1307 | 1336 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1309 | 1331 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 9.3E-10 | 1332 | 1360 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.18 | 1337 | 1363 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.926 | 1337 | 1368 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1339 | 1363 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1373 aa Download sequence Send to blast |
MSLQPPAVEP PEWLRTLPVA PEYHPTLAEF ADPIAYILRI EPEASRYGIC KIVPPLPRPP 60 EDDTFRRLQD AFAAAASSNG AGGDPSPTFP PTFPTRLQQV GLSARNRRAA SRRVWESGER 120 YTLEAFRAKA AEFEPPRHAA PPRNPTHLQL EALFWAACAS RPFSVEYGND MPGSAFASPD 180 ELPDAANATD VGETEWNMRV APRARGSLLR AMARDVAGVT TPMLYVAMLY SWFAWHVEDH 240 ELHSLNFLHF GKAKTWYGVP RDAMLAFEET VRVHGYADDL NAIMAFQTLN EKTTVLSPEV 300 LLSAGVPCCR LVQKAGEFVI TFPGAYHSGF SHGFNCGEAS NIATPHWLQV AKEAAIRRAS 360 TNCGPMVSHY QLLYELALSL RPREPKNFYS VPRSSRLRDK NKNEGDIMVK ENFVGSVAEN 420 NNLLSVLLDK NYCIIVPNTD FFVPSFPVAL ESEATVKQRF TAGPCSISQQ GAENMAVDHV 480 AVDKVTDIQD MSGSLYPCET SLVGCSNRKL YETKYGQRDA AALCLSTSEI QSRGIDKARS 540 HPAGGILDQG RLPCVQCGIL SFACVAIIQP REAAVQFIMS KECISSSAKQ GGIGASDDTS 600 NWIDQSHEIS PPPGPASGTD DNVKHTVSLA HVSDRCRELY ASNTDGCTSA LGLLASAYDS 660 SDSDDETTED VLKHSKKNDS VNQSMDTQIL ETSASYSSTV RCQKTNSHSH EEECEARATS 720 LMKPVSHNSR PISQSNRDTD IDQFIELGKS GTQCSGYLDL VDDLTTSVLK SSSDTCVSAA 780 KASMDPDVLT MLRYNKDSCR MHVFCLEHAL ETWTQLQQIG GANIMLLCHP EYPRAESAAK 840 VIAEELGIKH DWKDITFKEA TEEDVKKIRL ALQDEDAEPT GSDWAVKMGI NIYYSAKQSK 900 SPLYSKQIPY NSIIYKAFGQ ENPDSLTDYG CQKSGSTKKK VAGWWCGKVW MSNQVHPLLA 960 REREEQNSSV VYGKAMFTTI SHGKVQDEAS TRCNTSNRTP SKRTSRRKKG VSAEKSKPKN 1020 KRSTASDEAS MHCSGLGMNS GVIHDQTENS DDYDKHGNGD EIEEGTNPQK YQQRKLQNVT 1080 RKSSSKKRKD EKRTDSFHEL YDEDNGVDYW LNMGSGHDAT LGNSQQQSPD PVKVKSGGKL 1140 QGKRKSSKYK SNDDLLNEEN KLQKMNKKSS SKKQKNDKIN RQLQEDQTED DHMDHLVDVA 1200 VADEVTLDNE DKITEDKIDD VKVKSRGKSQ NGKRKGSKHQ ASDGLRAGNK VAKFPCDIEG 1260 CDMSFSTQQD LLLHKRDICP VKGCKKKFFC HKYLLQHRKV HIDERPLKCT WKGCKKAFKW 1320 PWARTEHMRV HTGVRPYECQ EPGCGQTFRF VSDFSRHKRK TGHSSDKRRK NST |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6ip0_A | 2e-69 | 12 | 380 | 7 | 353 | Transcription factor jumonji (Jmj) family protein |
| 6ip4_A | 2e-69 | 12 | 380 | 7 | 353 | Arabidopsis JMJ13 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
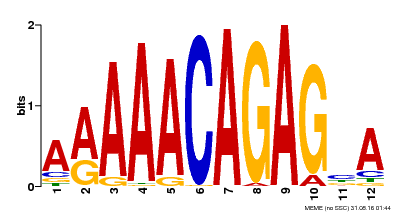
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK065251 | 0.0 | AK065251.1 Oryza sativa Japonica Group cDNA clone:J013002J08, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015620626.1 | 0.0 | lysine-specific demethylase REF6 | ||||
| TrEMBL | A0A0E0BRB6 | 0.0 | A0A0E0BRB6_9ORYZ; Uncharacterized protein | ||||
| STRING | OGLUM12G09840.1 | 0.0 | (Oryza glumipatula) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6718 | 30 | 35 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 1e-153 | relative of early flowering 6 | ||||



