 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OMERI01G39560.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 787aa MW: 88348.4 Da PI: 6.6646 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 69.7 | 3.9e-22 | 129 | 230 | 1 | 99 |
EEEE-..-HHHHTT-EE--HHH.HTT.......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SS CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh.......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgr 87
f+k+lt sd++++g +++ +++a+e+ +++++ ++l+ +d+++ W++++i+r++++r++l++GW+ Fv++++L +gD ++F +
OMERI01G39560.2 129 FCKTLTASDTSTHGGFSVLRRHADEClppldmtQSPPT--QELVAKDLHSMDWRFRHIFRGQPRRHLLQSGWSVFVSSKRLVAGDAFIFL--RG 218
99*****************************7555544..58************************************************..44 PP
SEE..EEEEE-S CS
B3 88 sefelvvkvfrk 99
+++el+v+v+r+
OMERI01G39560.2 219 ENGELRVGVRRA 230
9999*****996 PP
| |||||||
| 2 | Auxin_resp | 113.6 | 1.8e-37 | 255 | 336 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalkvkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
a+ha++tks+F+v+Y+Pr+s+seF+++++++++++k+++svGmRf+m+fe+e+++e+r++Gt++g ++ldp Wp+S WrsLk
OMERI01G39560.2 255 AWHAINTKSMFTVYYKPRTSPSEFIIPYDQYMESVKNNYSVGMRFRMRFEGEEAPEQRFTGTIIGSENLDP-VWPESSWRSLK 336
89*********************************************************************.9********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 8.24E-40 | 121 | 258 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 3.2E-39 | 122 | 244 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 5.10E-19 | 127 | 229 | No hit | No description |
| PROSITE profile | PS50863 | 11.265 | 129 | 231 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 1.5E-21 | 129 | 231 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 7.8E-20 | 129 | 230 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 5.7E-34 | 255 | 336 | IPR010525 | Auxin response factor |
| Pfam | PF02309 | 5.0E-14 | 622 | 726 | IPR033389 | AUX/IAA domain |
| SuperFamily | SSF54277 | 4.37E-13 | 637 | 718 | No hit | No description |
| PROSITE profile | PS51745 | 26.238 | 641 | 734 | IPR000270 | PB1 domain |
| Gene3D | G3DSA:3.10.20.240 | 9.5E-4 | 659 | 714 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0008285 | Biological Process | negative regulation of cell proliferation | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009911 | Biological Process | positive regulation of flower development | ||||
| GO:0010047 | Biological Process | fruit dehiscence | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0010227 | Biological Process | floral organ abscission | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0048481 | Biological Process | plant ovule development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 787 aa Download sequence Send to blast |
MPPAAMAPPP PPQGSSTGDP LYDELWHACA GPLVTVPRVG DLVFYFPQGH IEQVEASMNQ 60 VADSQMRLYD LPSKLLCRVL NVELKAEQDT DEVYAQVMLM PEPEQNEMAV EKTTPTSGPV 120 QARPPVRSFC KTLTASDTST HGGFSVLRRH ADECLPPLDM TQSPPTQELV AKDLHSMDWR 180 FRHIFRGQPR RHLLQSGWSV FVSSKRLVAG DAFIFLRGEN GELRVGVRRA MRQLSNVPSS 240 VISSQSMHLG VLATAWHAIN TKSMFTVYYK PRTSPSEFII PYDQYMESVK NNYSVGMRFR 300 MRFEGEEAPE QRFTGTIIGS ENLDPVWPES SWRSLKVRWD EPSTIPRPDR VSPWKIEPAS 360 SPPVNPLPLS RVKRPRPNAP PASPESPILM KEAATKVDTD PAQAQRSQNG TVLQGQEQMT 420 LRSNLTESND SDVTAHKPMM WSPSPNAAKA HPLTFQQRPP MDNWMQLGRR ETDFKDPSLT 480 VESSTQMHTD SKELHFWNGQ STTVYGNSRD QPQNFRFEQN SSSWLNQSFA RPEQPRVIRP 540 HASIAPVELE KTEGSGFKIF GFKVDTTNAP NNHLSSPMAA THEPMLQTPS SLNQLQPVQT 600 DCIPEVSVST AGMATENEKS GQQAQQSSKD VQSKTQVAST RSCTKVHKQG VALGRSVDLS 660 KFSNYDELKA ELDKMFEFDG ELVSSNKNWQ IVYTDNEGDM MLVGDDPWEE FCSIVRKIYI 720 YTKEEVQKMN SKSNAPRKED SSENEKGSVK RDDTRGSRFI IIAIAPLNSP QLSDVNIYLR 780 SQPPSQK |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldv_A | 1e-170 | 14 | 361 | 12 | 357 | Auxin response factor 1 |
| 4ldw_A | 1e-170 | 14 | 361 | 12 | 357 | Auxin response factor 1 |
| 4ldw_B | 1e-170 | 14 | 361 | 12 | 357 | Auxin response factor 1 |
| 4ldx_A | 1e-170 | 14 | 361 | 12 | 357 | Auxin response factor 1 |
| 4ldx_B | 1e-170 | 14 | 361 | 12 | 357 | Auxin response factor 1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 372 | 376 | KRPRP |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00574 | DAP | Transfer from AT5G62000 | Download |
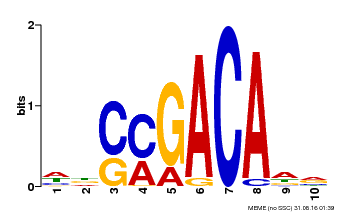
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK072309 | 0.0 | AK072309.1 Oryza sativa Japonica Group cDNA clone:J023022C23, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015650953.1 | 0.0 | auxin response factor 4 | ||||
| Swissprot | Q5JK20 | 0.0 | ARFD_ORYSJ; Auxin response factor 4 | ||||
| TrEMBL | A0A0E0CCH1 | 0.0 | A0A0E0CCH1_9ORYZ; Auxin response factor | ||||
| STRING | OMERI01G39070.1 | 0.0 | (Oryza meridionalis) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G62000.3 | 0.0 | auxin response factor 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



