 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OMERI02G00590.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 941aa MW: 106169 Da PI: 7.9963 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 17.3 | 1.3e-05 | 632 | 654 | 1 | 23 |
EEETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCpdCgksFsrksnLkrHirtH 23
++C+ Cg sF + +Lk+H+++H
OMERI02G00590.1 632 HTCQECGASFQKPAHLKQHMQSH 654
79*******************99 PP
| |||||||
| 2 | zf-C2H2 | 20.5 | 1.3e-06 | 660 | 684 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
+ Cp dC+ s+ rk++L+rH+ +H
OMERI02G00590.1 660 FICPleDCPFSYIRKDHLNRHMLKH 684
78*********************99 PP
| |||||||
| 3 | zf-C2H2 | 12.9 | 0.00034 | 689 | 714 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
++C+ C++ Fs k n++rH + H
OMERI02G00590.1 689 FTCSmdGCDRKFSIKANMQRHVKEiH 714
89********************9988 PP
| |||||||
| 4 | zf-C2H2 | 15.9 | 3.8e-05 | 726 | 750 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
++C+ C+k+F+ s Lk+H +H
OMERI02G00590.1 726 FVCKeeGCNKVFKYASRLKKHEESH 750
89*******************9988 PP
| |||||||
| 5 | zf-C2H2 | 16.4 | 2.6e-05 | 817 | 841 | 2 | 23 |
EET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 2 kCp..dCgksFsrksnLkrHirt.H 23
kC+ C sFs+ksnL++Hi+ H
OMERI02G00590.1 817 KCSfeGCECSFSNKSNLTKHIKAsH 841
688889***************9888 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.25.40.10 | 1.7E-8 | 80 | 218 | IPR011990 | Tetratricopeptide-like helical domain |
| PROSITE profile | PS51375 | 8.703 | 114 | 148 | IPR002885 | Pentatricopeptide repeat |
| PROSITE profile | PS51375 | 9.701 | 184 | 218 | IPR002885 | Pentatricopeptide repeat |
| TIGRFAMs | TIGR00756 | 2.3E-4 | 186 | 214 | IPR002885 | Pentatricopeptide repeat |
| Pfam | PF01535 | 1.7E-4 | 186 | 213 | IPR002885 | Pentatricopeptide repeat |
| Pfam | PF01535 | 0.59 | 217 | 244 | IPR002885 | Pentatricopeptide repeat |
| PROSITE profile | PS51375 | 10.073 | 246 | 280 | IPR002885 | Pentatricopeptide repeat |
| TIGRFAMs | TIGR00756 | 1.3E-4 | 248 | 281 | IPR002885 | Pentatricopeptide repeat |
| Pfam | PF01535 | 1.8E-4 | 248 | 277 | IPR002885 | Pentatricopeptide repeat |
| PROSITE profile | PS51375 | 7.903 | 316 | 346 | IPR002885 | Pentatricopeptide repeat |
| Gene3D | G3DSA:1.25.40.10 | 1.7E-8 | 317 | 347 | IPR011990 | Tetratricopeptide-like helical domain |
| Pfam | PF01535 | 0.032 | 321 | 345 | IPR002885 | Pentatricopeptide repeat |
| Pfam | PF13041 | 1.4E-10 | 346 | 394 | IPR002885 | Pentatricopeptide repeat |
| PROSITE profile | PS51375 | 11.641 | 347 | 381 | IPR002885 | Pentatricopeptide repeat |
| TIGRFAMs | TIGR00756 | 5.1E-6 | 349 | 382 | IPR002885 | Pentatricopeptide repeat |
| PROSITE profile | PS51375 | 9.219 | 382 | 412 | IPR002885 | Pentatricopeptide repeat |
| TIGRFAMs | TIGR00756 | 0.0011 | 384 | 411 | IPR002885 | Pentatricopeptide repeat |
| PROSITE profile | PS51375 | 7.048 | 418 | 448 | IPR002885 | Pentatricopeptide repeat |
| Pfam | PF01535 | 0.014 | 421 | 445 | IPR002885 | Pentatricopeptide repeat |
| Gene3D | G3DSA:1.25.40.10 | 1.7E-8 | 448 | 513 | IPR011990 | Tetratricopeptide-like helical domain |
| PROSITE profile | PS51375 | 5.579 | 450 | 480 | IPR002885 | Pentatricopeptide repeat |
| PROSITE profile | PS51375 | 7.969 | 484 | 518 | IPR002885 | Pentatricopeptide repeat |
| Pfam | PF01535 | 0.67 | 493 | 515 | IPR002885 | Pentatricopeptide repeat |
| SMART | SM00355 | 24 | 586 | 608 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 2.8E-5 | 629 | 651 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0017 | 632 | 654 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 13.547 | 632 | 659 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 634 | 654 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 9.3E-11 | 644 | 686 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.7E-11 | 652 | 686 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 8.808 | 660 | 689 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 5.5E-4 | 660 | 684 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 662 | 684 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 7.02E-8 | 670 | 720 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 3.1E-8 | 687 | 716 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 5.6E-4 | 689 | 714 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.572 | 689 | 719 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 691 | 714 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 5.5E-7 | 721 | 750 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0019 | 726 | 750 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.305 | 726 | 750 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 728 | 750 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 5.7 | 758 | 783 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 760 | 783 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 15 | 786 | 807 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 1.1E-5 | 798 | 838 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 1.36E-7 | 799 | 853 | No hit | No description |
| SMART | SM00355 | 0.002 | 816 | 841 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 11.365 | 816 | 846 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 818 | 841 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 5.8 | 847 | 873 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 849 | 873 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| GO:0008097 | Molecular Function | 5S rRNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0080084 | Molecular Function | 5S rDNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 941 aa Download sequence Send to blast |
MKARRNQEPH SSLPAAAAPS SSRRSAGNHT SSQPRSPPVQ THQRFQGGMN RSVCHHLLTQ 60 CKTIRELQRI HAQALAHGLH PNHQSISCKI FRSYAEFGRP AEAGRLFDEI PHPDIISFTS 120 LMSLHLKLDH HRKAISVFSH AIASGHRPDG FAAVGALSAS GGLGDQCIGS AVHGLIFRCG 180 LDFELVLCNA LVDMYCRCGK FEPARAVFDR MLVKDEVTWG SMLYGHMKCA GVDSALSFFY 240 QMPMKSTVSW TALITGHVQD KQPIQALELF GRMLLQGHRP THITIVGVLS ACADIGALDL 300 GRAIHGYGSK SNATTNIIVT NALMDMYAKS GSIASAFSVF EEVQMKDAFT WTTMISSFTV 360 QGNGRKAVEL FWDMLRSGIL PNSVTFVSVL SACSHAGLIQ EGRELFDKMR EVYHIDPRLE 420 HYGCMVDLLG RSGLLEEAEA LIDHMDVEPD IVIWRSLLSA CLAHGNDRLA EIAGKEIIKR 480 EPGDDGVYVL LWNMYASSNR WKEALDVRKQ MLSRKIYKKP GCSWIEVDGV VHEFLVEDKT 540 HDARREIYGT LEIMARHLKM DPMCSGDDID GDTRVEATQH RDIRRYKCEF CTVVRSKKCL 600 IRAHMVAHHK EELDKSEIYK SNGEKVVHEG DHTCQECGAS FQKPAHLKQH MQSHSDERSF 660 ICPLEDCPFS YIRKDHLNRH MLKHQGKLFT CSMDGCDRKF SIKANMQRHV KEIHEDENAT 720 KSNQQFVCKE EGCNKVFKYA SRLKKHEESH VKLDYVEVVC CEPGCMKTFT NVECLRAHNQ 780 ACHQYVQCDI CGEKHLKKNI KRHLRAHEEV PSTERIKCSF EGCECSFSNK SNLTKHIKAS 840 HDQVKPFACR FTGCEKVFPY KHVRDNHEKS SAHVYTQANF TEMDEQLLSC PRGGRKRKAV 900 TVETLTRKRV TMHGDASSLD NGTEYLRWLL SGGDDDSSQT H |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5iww_D | 1e-55 | 68 | 516 | 10 | 333 | PLS9-PPR |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00229 | DAP | Transfer from AT1G72050 | Download |
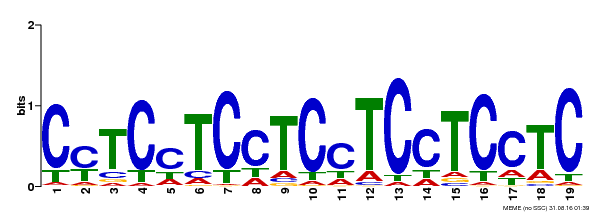
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | CP012610 | 0.0 | CP012610.1 Oryza sativa Indica Group cultivar RP Bio-226 chromosome 2 sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025878917.1 | 0.0 | pentatricopeptide repeat-containing protein At2g22410, mitochondrial | ||||
| TrEMBL | A0A0E0CDW1 | 0.0 | A0A0E0CDW1_9ORYZ; Uncharacterized protein | ||||
| STRING | OMERI02G00620.1 | 0.0 | (Oryza meridionalis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2382 | 38 | 82 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72050.2 | 1e-117 | transcription factor IIIA | ||||



