 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OMERI02G02950.5 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 657aa MW: 73894.1 Da PI: 7.1487 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 114.7 | 1.4e-35 | 1 | 124 | 29 | 142 |
DUF822 29 iyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpl..eeaeaagssasaspesslq.sslkssalaspvesysaspksssf 119
+++GLR++Gn++lp+raD+n+Vl+AL+r+AGw+v++DGtt+r++s+pl +++ g ++ +s+e+++ s+l+s+a+ +p++s++++++++++
OMERI02G02950.5 1 MLSGLRQHGNFPLPARADMNDVLAALARAAGWTVHPDGTTFRASSQPLhtPTPQLPGIFHVNSVETPSFtSVLNSYAIGTPLDSQASMLQTDDS 94
79*********************************************998999**************99899********************** PP
DUF822 120 pspssldsislasa.......asllpvlsv 142
+spsslds+ +a++ ++ +v+s
OMERI02G02950.5 95 LSPSSLDSVVVAEQsiknekyGNSDSVSSL 124
***********9987766655444444444 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 5.1E-33 | 1 | 127 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| Gene3D | G3DSA:3.20.20.80 | 2.8E-152 | 145 | 500 | IPR013781 | Glycoside hydrolase, catalytic domain |
| SuperFamily | SSF51445 | 3.93E-151 | 145 | 565 | IPR017853 | Glycoside hydrolase superfamily |
| Pfam | PF01373 | 1.1E-74 | 170 | 498 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.3E-42 | 183 | 197 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.3E-42 | 204 | 222 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.3E-42 | 317 | 339 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.3E-42 | 390 | 409 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.3E-42 | 424 | 440 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.3E-42 | 441 | 452 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.3E-42 | 459 | 482 | IPR001554 | Glycoside hydrolase, family 14 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000272 | Biological Process | polysaccharide catabolic process | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0048831 | Biological Process | regulation of shoot system development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0016161 | Molecular Function | beta-amylase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 657 aa Download sequence Send to blast |
MLSGLRQHGN FPLPARADMN DVLAALARAA GWTVHPDGTT FRASSQPLHT PTPQLPGIFH 60 VNSVETPSFT SVLNSYAIGT PLDSQASMLQ TDDSLSPSSL DSVVVAEQSI KNEKYGNSDS 120 VSSLNCLENH QLTRASAALA GDYTRTPYIP VYASLPMGII NSHCQLIDPE GIRAELMHLK 180 SLNVDGVIVD CWWGIVEAWI PHKYEWSGYR DLFGIIKEFK LKVQVVLSFH GSGETGSGGV 240 SLPKWVMEIA QENQDVFFTD REGRRNMECL SWGIDKERVL RGRTGIEAYF DFMRSFHMEF 300 RNLTEEGLIS AIEIGLGVSG ELKYPSCPER MGWRYPGIGE FQCYDRYMQK NLRQAALSRG 360 HLFWARGPDN AGYYNSRPHE TGFFCDGGDY DSYYGRFFLN WYSGILIDHV DQVLSLATLA 420 FDGVETVVKI PSIYWWYRTA SHAAELTAGF YNPTNRDGYS PVFRMLKKHS VILKFVCYGP 480 EFTIQENNEA FADPEGLTWQ RDVCFSELGN FVKCMHDGPL IKIVVRVRII THQSRSSVLD 540 VLMQRLTVLC IRGVSTYPIK IIRSRYLTTI IAERDRSPTE ADSVRHGPTW LALAAAELAM 600 ARRAALDLGG ARLRLLPPRP PPHRRRPPPP SLRFPPPKST ESQRRWDVDG TTTLIDP |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wdq_A | 1e-111 | 148 | 477 | 11 | 343 | Beta-amylase |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00543 | DAP | Transfer from AT5G45300 | Download |
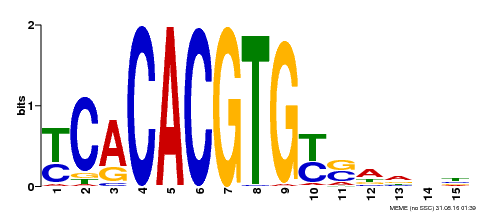
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK120049 | 0.0 | AK120049.1 Oryza sativa Japonica Group cDNA clone:J013000A07, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015622759.1 | 0.0 | beta-amylase 8 | ||||
| Swissprot | Q9FH80 | 0.0 | BAM8_ARATH; Beta-amylase 8 | ||||
| TrEMBL | A0A0E0CEX4 | 0.0 | A0A0E0CEX4_9ORYZ; Beta-amylase | ||||
| STRING | ONIVA02G02260.2 | 0.0 | (Oryza nivara) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45300.2 | 0.0 | beta-amylase 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



