 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OMERI02G20310.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | ARF | ||||||||
| Protein Properties | Length: 680aa MW: 74922.2 Da PI: 6.3001 | ||||||||
| Description | ARF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | B3 | 73.4 | 2.7e-23 | 130 | 230 | 1 | 98 |
EEEE-..-HHHHTT-EE--HHH.HTT.......---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SS CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh.......ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgr 87
f+k+lt sd++++g +++ +++aee+ ++++ + +l+ +d++g++W +++i+r++++r++lt+GW+ Fv++++L +gD ++F +
OMERI02G20310.1 130 FCKTLTASDTSTHGGFSVLRRHAEEClppldmtQNPPWQ--ELVARDLHGNEWHFRHIFRGQPRRHLLTTGWSVFVSSKRLVAGDAFIFL--RG 219
99****************************986555545..8************************************************..44 PP
SEE..EEEEE- CS
B3 88 sefelvvkvfr 98
+++el+v+v+r
OMERI02G20310.1 220 ENGELRVGVRR 230
9999*****97 PP
| |||||||
| 2 | Auxin_resp | 111.4 | 9.1e-37 | 256 | 338 | 1 | 83 |
Auxin_resp 1 aahaastksvFevvYnPrastseFvvkvekvekalkvkvsvGmRfkmafetedsserrlsGtvvgvsdldpvrWpnSkWrsLk 83
a+ha+st++ F+v+Y+Pr+s+seFvv+++k+++a ++k+svGmRfkm+fe+++++err+sGt++gv ++++ W nS WrsLk
OMERI02G20310.1 256 ASHAISTGTLFSVFYKPRTSQSEFVVSANKYLEAKNSKISVGMRFKMRFEGDEAPERRFSGTIIGVGSMSTSPWANSDWRSLK 338
789******************************************************************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101936 | 4.58E-44 | 119 | 259 | IPR015300 | DNA-binding pseudobarrel domain |
| Gene3D | G3DSA:2.40.330.10 | 4.6E-41 | 125 | 245 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 7.80E-22 | 128 | 230 | No hit | No description |
| SMART | SM01019 | 1.5E-22 | 130 | 232 | IPR003340 | B3 DNA binding domain |
| Pfam | PF02362 | 5.4E-21 | 130 | 230 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 11.462 | 130 | 232 | IPR003340 | B3 DNA binding domain |
| Pfam | PF06507 | 1.9E-34 | 256 | 338 | IPR010525 | Auxin response factor |
| Pfam | PF02309 | 5.1E-13 | 537 | 641 | IPR033389 | AUX/IAA domain |
| PROSITE profile | PS51745 | 25.319 | 550 | 643 | IPR000270 | PB1 domain |
| SuperFamily | SSF54277 | 6.54E-12 | 552 | 629 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 680 aa Download sequence Send to blast |
MAGSVAAAAA AAASGGGTGS SCDALYRELW HACAGPLVTV PRQGELVYYF PQGHMEQLEA 60 STDQQLDQHL PLFNLPSKIL CKVVNVELRA ETDSDEVYAQ IMLQPEADQN ELTSPKPEPH 120 EPEKCNVHSF CKTLTASDTS THGGFSVLRR HAEECLPPLD MTQNPPWQEL VARDLHGNEW 180 HFRHIFRGQP RRHLLTTGWS VFVSSKRLVA GDAFIFLRGE NGELRVGVRR LMRQLNNMPS 240 SVISSHSMHL GVLATASHAI STGTLFSVFY KPRTSQSEFV VSANKYLEAK NSKISVGMRF 300 KMRFEGDEAP ERRFSGTIIG VGSMSTSPWA NSDWRSLKVQ WDEPSVVPRP DRVSPWELEP 360 LAASNSQSSP QPPARNKRAR PPASNSIAPE LPPVFGLWKS SAESTQGFSF SGLQRTQELY 420 PSSPNLIFST SLNVGFSTKN EPSALSNKHF YWPMRETRAD SYSASISKVP SEKKQEPSSA 480 GCRLFGIEIS SAVEATSPLA AVSGVGQDQP AASVDAESDQ LSQPSHANKS DAPAASSEPS 540 PHETQSRQVR SCTKVIMQGM AVGRAVDLTR LHGYDDLRCK LEEMFDIQGE LSASLKKWKV 600 VYTDDEDDMM LVGDDPWPEF CSMVKRIYIY TYEEAKQLTP KLKLPIIGDA IKPNPNKQSP 660 ESDMPHSDLD STAPVTDKDC |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4ldv_A | 0.0 | 8 | 368 | 2 | 361 | Auxin response factor 1 |
| 4ldw_A | 0.0 | 8 | 368 | 2 | 361 | Auxin response factor 1 |
| 4ldw_B | 0.0 | 8 | 368 | 2 | 361 | Auxin response factor 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Auxin response factors (ARFs) are transcriptional factors that bind specifically to the DNA sequence 5'-TGTCTC-3' found in the auxin-responsive promoter elements (AuxREs). | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00097 | PBM | Transfer from AT1G59750 | Download |
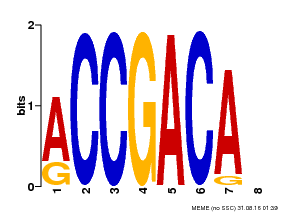
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK070026 | 0.0 | AK070026.1 Oryza sativa Japonica Group cDNA clone:J023036G16, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015625501.1 | 0.0 | auxin response factor 7 | ||||
| Swissprot | Q6YVY0 | 0.0 | ARFG_ORYSJ; Auxin response factor 7 | ||||
| TrEMBL | A0A0E0CM01 | 0.0 | A0A0E0CM01_9ORYZ; Auxin response factor | ||||
| STRING | OMERI02G19450.1 | 0.0 | (Oryza meridionalis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1082 | 36 | 116 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G59750.4 | 0.0 | auxin response factor 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



