 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OMERI03G34570.3 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 604aa MW: 66284.4 Da PI: 10.5827 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 24.6 | 4.5e-08 | 144 | 188 | 5 | 54 |
HHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHH CS
HLH 5 hnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksL 54
h+++Er RR+ri +++ L+el+P+ K + a++L + +Y+k L
OMERI03G34570.3 144 HSIAERLRRERIAERMRALQELVPNT-----NKTDRAAMLDEILDYVKFL 188
99***********************7.....5***************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 1.96E-14 | 137 | 198 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 7.81E-8 | 137 | 193 | No hit | No description |
| PROSITE profile | PS50888 | 15.77 | 139 | 188 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 8.8E-14 | 141 | 199 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.9E-5 | 144 | 188 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.2E-9 | 145 | 194 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF10153 | 1.9E-26 | 325 | 431 | IPR019310 | rRNA-processing protein Efg1 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006364 | Biological Process | rRNA processing | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 604 aa Download sequence Send to blast |
MAGQQPQQQG PPEDDFFDQF FSLTSSFPGA APGGRAAGDQ PFSLALSLDA AAAAEASGSG 60 KRLGVGDDAE GGGSKADRET VQLTGLFPPV FGGGGVQPPN LRPTPPTQVF HPQQSKQGGA 120 AVGPQPPAPR PKVRARRGQA TDPHSIAERL RRERIAERMR ALQELVPNTN KTDRAAMLDE 180 ILDYVKFLRL QVKVLSMSRL GGAGAVAQLV ADIPLSVKGE ASDSGGNQQI WEKWSTDGTE 240 RQVAKLMEED IGAAMQFLQS KALCMMPISL AMAIYDTQQT QDGQPAMAHG RAAAVRRPKS 300 SSASSAGAAA ERKRKRAAAA KTVSLKNQIR STERLLRKDL PNDIRVAQEK KLEELKRQQE 360 LQNQLAIQRT VQLRDRKIKF FERRKIERMI RRLEKQQRSN ADDASNKLSK LKEDLEYVRF 420 FPKNEKYVSL FSGGNTPDML EKRNKWRKQI KENLMAAAEN GKDLEETASD DDTVDVSDDD 480 FFMSGSSSDE EADDEWTDKS AKEPASSASG KAASGMSSDE KNQRQRDARV LMPPPRSLAP 540 NRTRPGVKHV LSSSSNTSNS TSGGTFKNRR AANQPGDHNS NLSSNSDAHK PRRKRRHRKK 600 KKLA |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 148 | 153 | RLRRER |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00659 | PBM | Transfer from Pp3c14_15890 | Download |
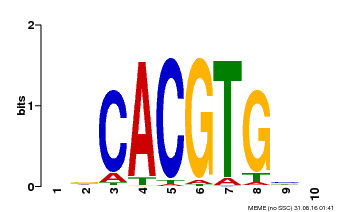
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK061648 | 0.0 | AK061648.1 Oryza sativa Japonica Group cDNA clone:001-036-B12, full insert sequence. | |||
| GenBank | AK098876 | 0.0 | AK098876.1 Oryza sativa Japonica Group cDNA clone:J013001D18, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015628282.1 | 0.0 | rRNA-processing protein efg1 | ||||
| TrEMBL | A0A0E0D877 | 0.0 | A0A0E0D877_9ORYZ; Uncharacterized protein | ||||
| STRING | OS03T0797700-01 | 0.0 | (Oryza sativa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6666 | 2 | 2 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G02590.3 | 3e-50 | bHLH family protein | ||||



