 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OMERI04G12590.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 423aa MW: 44853.9 Da PI: 7.7292 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 41.5 | 3.4e-13 | 161 | 242 | 2 | 86 |
trihelix 2 WtkqevlaLiearremeerlrrgklkkplWeevskkm.rergfer...spkqCkekwenlnkrykkikegekkrtsessstcpyfdqle 86
W++ + aLi+a e+ +l+rg+l+ p+W+ev++++ +++g+ + s +qCk++++ l+k+yk +++ + ++++f +l+
OMERI04G12590.1 161 WSEGATAALIDAWGERFVALGRGSLRHPQWQEVADAVsSREGYAKapkSDVQCKNRIDTLKKKYKIERAKPAS-------SWQFFGRLD 242
*************************************87888866222667**************99988876.......488888876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF13837 | 1.9E-22 | 159 | 244 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009793 | Biological Process | embryo development ending in seed dormancy | ||||
| GO:0010431 | Biological Process | seed maturation | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 423 aa Download sequence Send to blast |
MVGEVETGLF LAAGWPAGRS LPPILSRSRF RPRPDLTHHL LGAGPSIPPA WPKSTPSPRR 60 VTTGSPLASI PPPPHRRPQT RSDRSPNPAA AAAEMDDDDA AASASPSPSP SPVASALPVA 120 DPVTVAAGPP SGLLALALPI QKQQHAASPN PGGGGGREDA WSEGATAALI DAWGERFVAL 180 GRGSLRHPQW QEVADAVSSR EGYAKAPKSD VQCKNRIDTL KKKYKIERAK PASSWQFFGR 240 LDDLLAPTFN QKPGGNGGGG GGGAGVNGRN PVPAALRVGF PQRSRTPLMP APVSAVKRRA 300 PSPEPSASSE SSDGFPPERE PAFPPLPLPP PPNGKRSRAD EGRGGGGGDR AQGLRELAQA 360 IRRFGEAYER VETAKLEQSA EMERRRLDFA SELESQRVQF FLNTQMELSQ SDDEEDDEEE 420 ESQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription regulator that may repress the maturation program during early embryogenesis. {ECO:0000269|PubMed:21330492}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00356 | DAP | Transfer from AT3G14180 | Download |
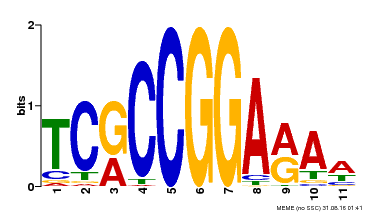
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AL731593 | 0.0 | AL731593.2 Oryza sativa genomic DNA, chromosome 4, BAC clone: OSJNBa0027P08, complete sequence. | |||
| GenBank | AP014960 | 0.0 | AP014960.1 Oryza sativa Japonica Group DNA, chromosome 4, cultivar: Nipponbare, complete sequence. | |||
| GenBank | CR855182 | 0.0 | CR855182.1 Oryza sativa genomic DNA, chromosome 4, BAC clone: OSIGBa0140O07, complete sequence. | |||
| GenBank | CT837970 | 0.0 | CT837970.1 Oryza sativa (indica cultivar-group) cDNA clone:OSIGCRA104F09, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015633896.1 | 1e-155 | trihelix transcription factor ASIL2 | ||||
| Swissprot | Q9LJG8 | 5e-38 | ASIL2_ARATH; Trihelix transcription factor ASIL2 | ||||
| TrEMBL | A0A0E0DET6 | 0.0 | A0A0E0DET6_9ORYZ; Uncharacterized protein | ||||
| STRING | OMERI04G11670.1 | 0.0 | (Oryza meridionalis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP2404 | 30 | 69 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G14180.1 | 2e-37 | sequence-specific DNA binding transcription factors | ||||



