 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | OMERI04G13990.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae; Oryzinae; Oryza
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 855aa MW: 92267.2 Da PI: 9.5571 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 83.4 | 2.9e-26 | 519 | 587 | 2 | 70 |
trihelix 2 WtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekk 70
W ++e+l+Li++rrem+++++++k++k+lWe++s++mre+gf rsp++C++kw+nl k++kk + ++++
OMERI04G13990.1 519 WVQDETLCLIALRREMDSHFNTSKSNKHLWEAISARMREQGFDRSPTMCTDKWRNLLKEFKKARSHARG 587
***************************************************************999876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF81383 | 4.19E-6 | 23 | 83 | IPR001810 | F-box domain |
| Gene3D | G3DSA:1.20.1280.50 | 6.1E-4 | 27 | 71 | No hit | No description |
| PROSITE profile | PS50090 | 8.293 | 511 | 575 | IPR017877 | Myb-like domain |
| Gene3D | G3DSA:1.10.10.60 | 9.3E-4 | 512 | 579 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.5E-5 | 515 | 577 | IPR001005 | SANT/Myb domain |
| CDD | cd12203 | 4.67E-29 | 517 | 582 | No hit | No description |
| Pfam | PF13837 | 1.4E-17 | 518 | 588 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 855 aa Download sequence Send to blast |
MSSSSSPRAP GDSSTRKRRR RLASGSGPRW ASLPEDLVDL VASRLLAGGD LLDYVRFRAV 60 CTSWRSATAS PRGRGVADRR FHPRRWMMLP EGHGLYPGHP SLRGYARFLN LDTGTLVRAR 120 IPLLRDGYVV IDPVDGLLLL LLDPDRNQEG AVRLLHPFTG DTAELPPLGT VLPHLGSRLL 180 DCPAPSRIRS LARVVCASVS CSATGAGAGA ITVLLALSVV SRVAFATSLD RQWSLSTYEC 240 VTLSSPIASH GKIYLMHTDR SCGEKIHQVL RIDPPAAAKD GSGSGSGAGR ALQEPKLVAT 300 IPARKLDHFQ GLVECGSEIL VLGCKNWSTS RISVFKLADL VLQRFMPIKS IGGHTLFIGE 360 RNISVSSKIL PTVKGDNLVY LNSGLVKYHL SSGSLSLAID NCSLYGRVPG PSSLVHYIYS 420 CCIRNRWPWP GTRAGPGRTS TASSPSPPGS PLLRSPTAAT TAPVTSPIAG FPFAHGATPR 480 APPRXXXXXX SSGEDGGHDS SSRAAASGGG GPKKRAETWV QDETLCLIAL RREMDSHFNT 540 SKSNKHLWEA ISARMREQGF DRSPTMCTDK WRNLLKEFKK ARSHARGGGG VGGGGVGGGA 600 GGGNCPAKMA CYKEIDDLLK RRGKPAGGGG AAVGSGAVKS PTVTSKIDSY LQFDKGFEDA 660 SIPFGPVEAS GRSLLSVEDR LEPDSHPLAL TADAVATNGV NPWNWRDTST NGGDNQVTFG 720 GRVILVKWGD YTKRIGIDGT ADAIKEAIKS AFGLRTRRAF WLEDEDEVVR SLDRDMPVGT 780 YTLHLDTGMT IKLYMFENDE VRTEDKTFYT EEDFRDFLSR RGWTLLREYS GYRIADTLDD 840 LRPGVIYEGM RSLGD |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2jmw_A | 8e-38 | 513 | 614 | 1 | 86 | DNA binding protein GT-1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 15 | 19 | RKRRR |
| 2 | 16 | 21 | KRRRRL |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00640 | PBM | Transfer from MDP0000164819 | Download |
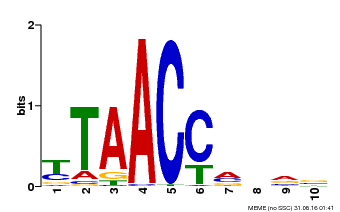
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK063256 | 0.0 | AK063256.1 Oryza sativa Japonica Group cDNA clone:001-112-H10, full insert sequence. | |||
| GenBank | AL606998 | 0.0 | AL606998.3 Oryza sativa genomic DNA, chromosome 4, BAC clone: OJ000223_09, complete sequence. | |||
| GenBank | AP014960 | 0.0 | AP014960.1 Oryza sativa Japonica Group DNA, chromosome 4, cultivar: Nipponbare, complete sequence. | |||
| GenBank | CP012612 | 0.0 | CP012612.1 Oryza sativa Indica Group cultivar RP Bio-226 chromosome 4 sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| TrEMBL | A0A0E0DFE3 | 0.0 | A0A0E0DFE3_9ORYZ; Uncharacterized protein | ||||
| STRING | ONIVA04G14730.1 | 0.0 | (Oryza nivara) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP5631 | 37 | 52 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G13450.1 | 1e-124 | Trihelix family protein | ||||



